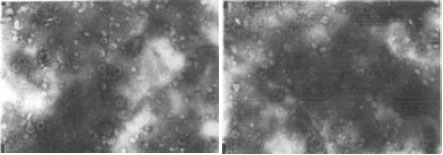
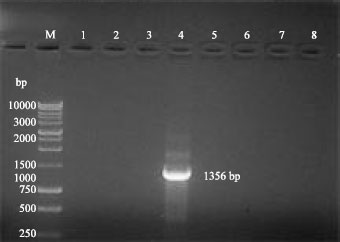
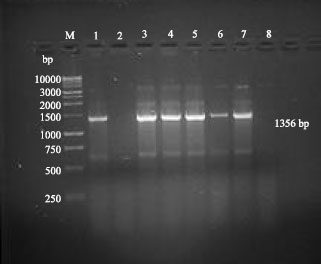
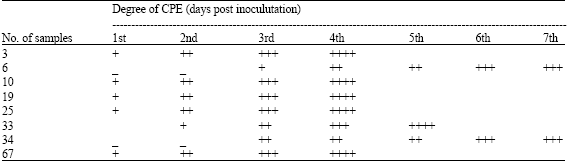
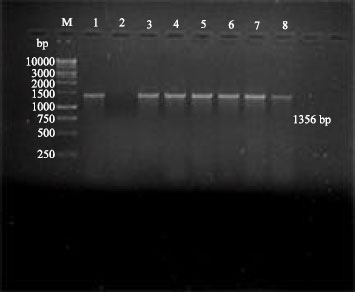
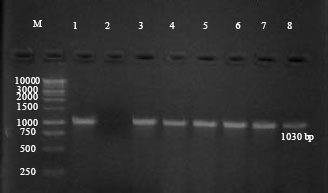
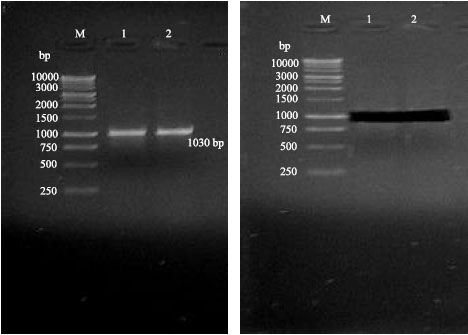
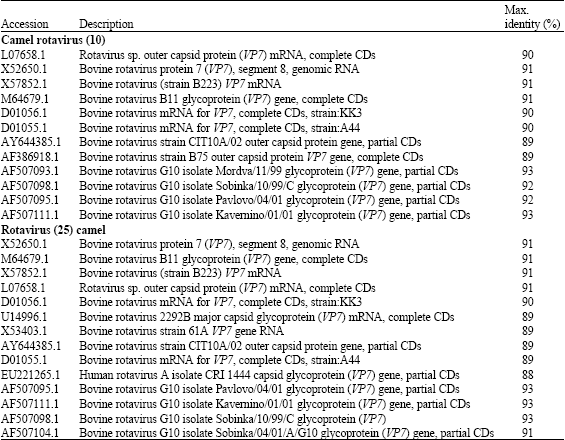
Research Article
Isolation and Antigenic and Molecular Characterization of G10 of Group A Rotavirus in Camel
Department of Virology, Animal Health Research Institue, Dokki, Giza, Egypt
H.A. Hussein
Department of Virology, Faculty of Veterinary Medicine, Cairo University, Giza, 12211, Egypt
I.M. El-Sabagh
Department of Virology, Faculty of Veterinary Medicine, Cairo University, Giza, 12211, Egypt
M.S. Saber
Department of Virology, Faculty of Veterinary Medicine, Cairo University, Giza, 12211, Egypt