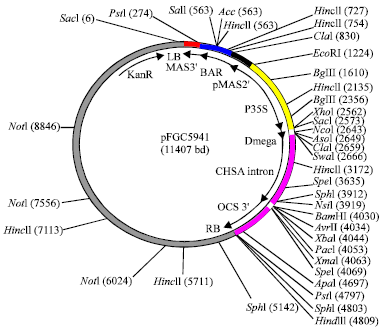
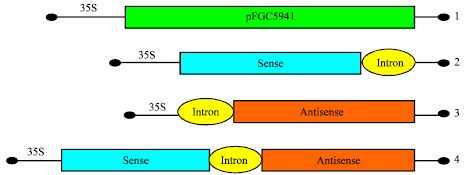
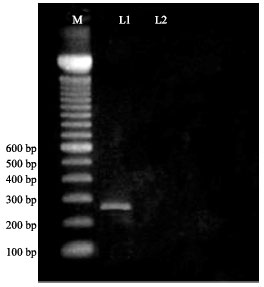
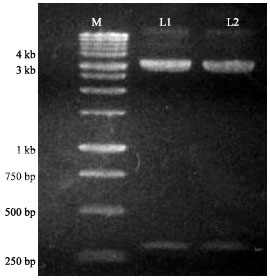
Research Article
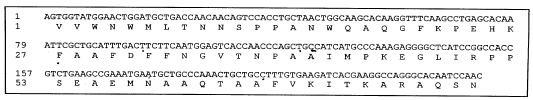
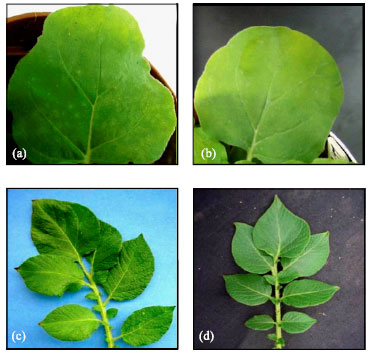
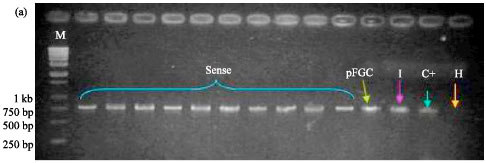
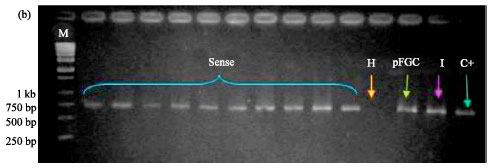
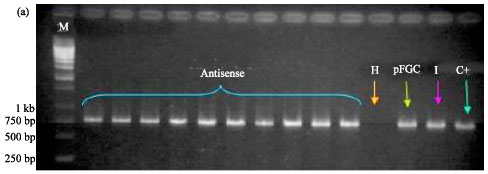
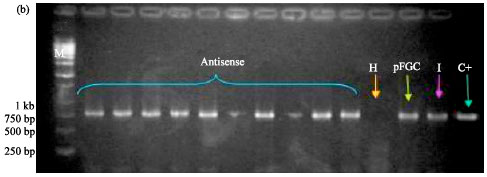
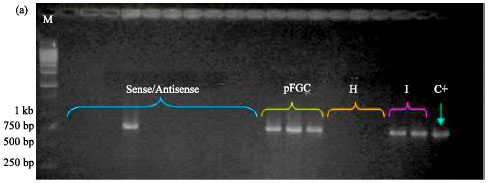
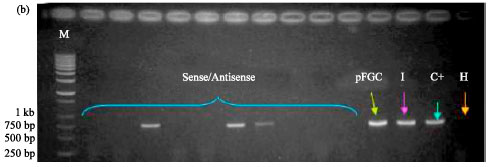
siRNA Silencing of PVX Coat Protein Gene Affects Accumulation of Viral RNA in Potato and Tobacco Plants
Plant Pathology Research Institute, Agriculture Research Center, Giza, Egypt
B.N. Barsoum
Department of Chemistry, Faculty of Science, Cairo University, Giza, Egypt
G.G. Mohamed
Department of Chemistry, Faculty of Science, Cairo University, Giza, Egypt
A.A. Rezk
Plant Pathology Research Institute, Agriculture Research Center, Giza, Egypt
A.E. Aboul-Ata
Plant Pathology Research Institute, Agriculture Research Center, Giza, Egypt
H.M. Mazyad
Plant Pathology Research Institute, Agriculture Research Center, Giza, Egypt