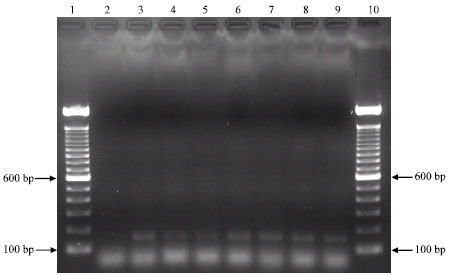
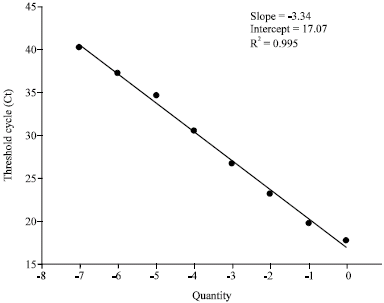
Research Article
A TaqManR RT-PCR Assay for the Detection of Feline calicivirus
Department of Veterinary Population Medicine, University of Minnesota, Saint Paul, MN 55108, USA
Ashok K. Tiwari
Department of Veterinary Population Medicine, University of Minnesota, Saint Paul, MN 55108, USA
Suchitra Sajja
Department of Veterinary Population Medicine, University of Minnesota, Saint Paul, MN 55108, USA
M. A. Ramakrishnan
Department of Veterinary Population Medicine, University of Minnesota, Saint Paul, MN 55108, USA
Kay S. Faaberg
Department of Veterinary and Biomedical Sciences, College of VeterinaiY Medicine, University of Minnesota, Saint Paul, MN 55108, USA
Sagar M. Goyal
Department of Veterinary Population Medicine, University of Minnesota, Saint Paul, MN 55108, USA