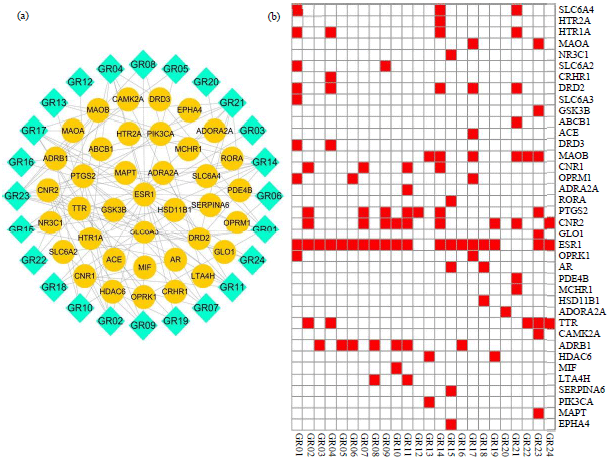
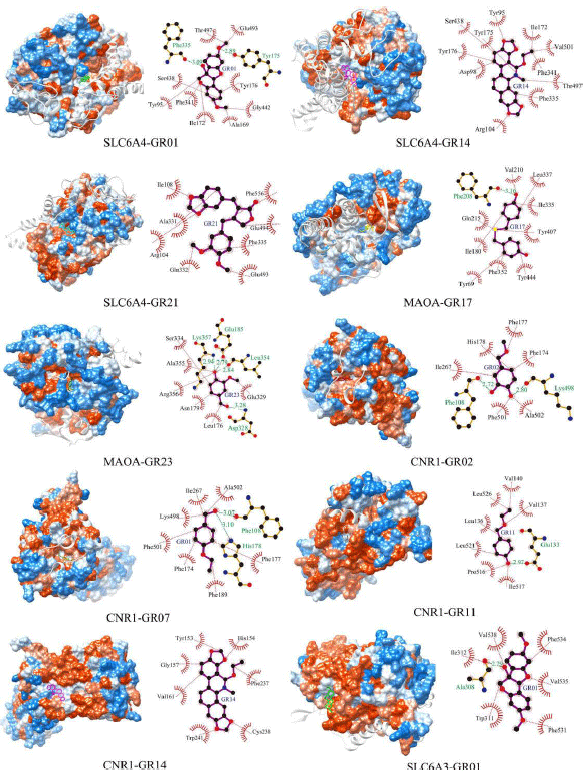
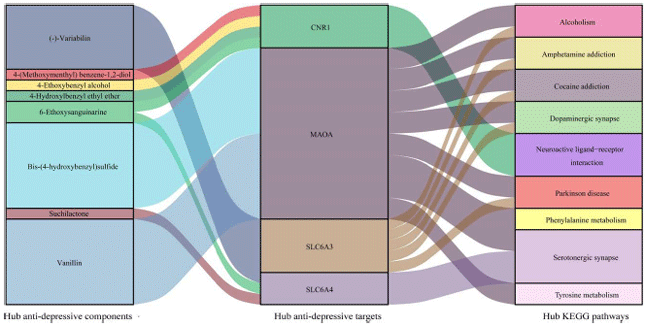
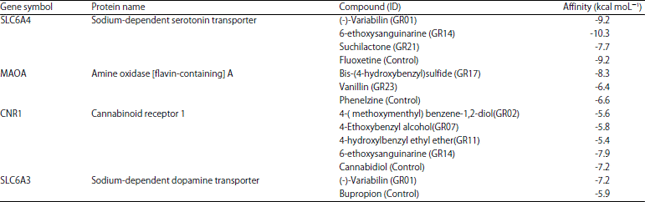
Research Article
Pharmacological Targets and Active Components of Gastrodiae rhizoma Against Depression: Findings of Network Pharmacology
Department of Pharmacy, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
LiveDNA: 86.33391
Cuicui Liu
Department of Clinical Laboratory, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
Jingjing Duan
Department of Pharmacy, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
Ting Zhou
Department of Pharmacy, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
Xiufeng Liu
Department of Pharmacy, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
Saihua Lu
Department of Pharmacy, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
Zhen Yang
Department of Central Laboratory, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
Feng Xu
Department of Pharmacy, Shanghai University of Medicine and Health Sciences Affiliated Sixth People's Hospital South Campus, Shanghai, People’s Republic of China
LiveDNA: 86.30347