Research Article
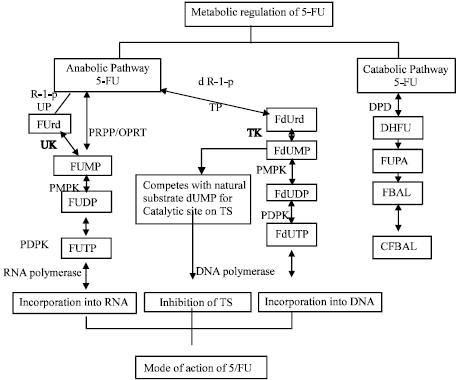
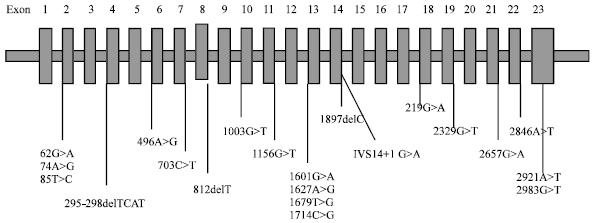
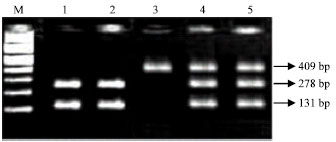
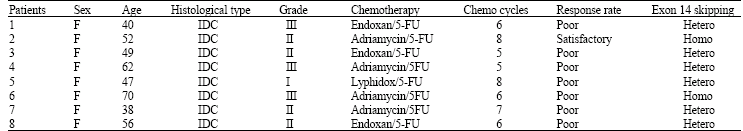
Possible Associations of Splice Site Mutation of Dihydropyrimidine Dehydrogenase (IVS14+1G>A) in Adverse Drug Reactions in Some Invasive Ductal Carcinoma Patients
Department of Genetics, Bhagawan Mahavir Medical Research Centre I C)- I - I Mahavir Marg, Hyderabad-500004, A.P., India
Sudha Murthy
Indo American Cancer Institute and Research, Centre Road No. 14: Banjara Hills Hyderabad-500034, A.P., India
Kaiser Jamil
School of Biotechnology (MGNIRSA), University ofMysore, Mysore, India