Research Article
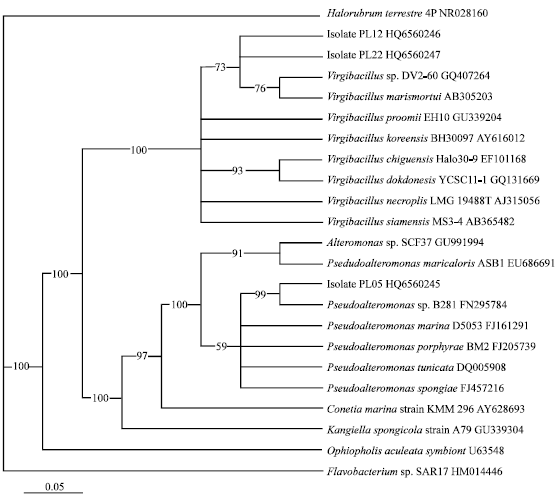
Screening of Multi-metal Resistances in a Bacterial Population Isolated from Coral Tissues of Central Java Coastal Waters, Indonesia
Department of Marine Science, FPK, Diponegoro University, Semarang, Indonesia
O.K. Radjasa
Department of Marine Science, FPK, Diponegoro University, Semarang, Indonesia
H.S. Utomo
Rice Research Station, Louisiana State University Ag Center, Rayne, LA, USA