ABSTRACT
A total of 8 samples of cow raw milks were collected from different areas of Khartoum state were analyzed for bacterial load. A total of 23 strains of lactic acid bacteria were isolated, out of which 12 strains were cocci and 11 strains were facultatively heterofermentative lactobacilli. Lactic acid bacteria were identified on the basis of phenotypic characters as Lactococcus lactis, Enterococcus faecalis, Enterococcus faecium, Enterococcus durans, Lactobacillus paracasei subsp. paracasei, Lactobacillus plantarum and Lactobacillus rhamnosus.
PDF Abstract XML References Citation
How to cite this article
DOI: 10.3923/ijds.2011.66.71
URL: https://scialert.net/abstract/?doi=ijds.2011.66.71
INTRODUCTION
In the history, milk played a major role as nutritional source and since 1900’s the start of golden era of industrial microbiology. It was also economically significant because larger quantity of milk was being processed daily in factories for the fermented food products. LAB were first isolated from milk (Carr et al., 2002; Metchnikoff, 1908; Sandine et al., 1972) and have since been found in such foods and fermented products as meat, milk products, vegetables, beverages and bakery products (Aukrust and Blom, 1992; Caplice and Fitzgerald, 1999; Harris et al., 1992; Gobbetti and Corsetti, 1997; Jay, 2000; Liu, 2003; Lonvaud-Funel, 2001; O’Sullivan et al., 2002). Lactic acid bacteria are widely distributed in the nature. They could be isolated from soils, waters, plants, silages, waste products and also from the intestinal tract of animals and humans (Axelsson, 1998). The genera Carnobacterium, Enterococcus, Lactobacillus, Lactococcus, Lactosphaera, Leuconostoc, Melissococcus, Oenococcus, Pediococcus, Streptococcus, Tetragenococcus, Vagococcus and Weissella are recognized as LAB (Jay, 2000). The objectives of this study were to collect a variety of milk samples from different areas of Khartoum state and to determine the predominant lactic acid bacteria groups (isolation and identification of microorganisms from cow's milk).
MATERIALS AND METHODS
Samples: The samples of cow's milk were collected from various areas of Khartoum state. They were obtained under good conditions from a healthy animal, to avoid any contamination which can influence the lactic flora. The samples were collected in sterile bottles and then transported quickly to the laboratory to be analyzed. The analysis was done at the laboratory of the Department of Food Microbiology, Food Research Centre, Khartoum North, Sudan. Sterilization, serial dilution and preparation of the media were done according to Harrigan and MacCance (1976).
Isolation of lactic acid bacteria from raw milk: Ten milliliter of milk sample were vigorously homogenized with 9 volumes of sterile diluents [0.1% (w/v) bacteriological peptone, 0.85% (w/v) [NaCl] and serial 10-fold dilutions (10-1 to 10-8) were prepared using the same diluents. One milliliter of these dilutions was pour-plated in the media for lactic acid bacteria, M17 (Terzaghi and Sandine, 1975) and MRS (De Man et al., 1960) adjusted to pH 5.5. After incubation anaerobically (BBL Gas pak plus Anaerobic System) at 37°C for 3 days, representative strains of lactic acid bacteria were obtained from M17 and MRS plates of highest sample dilutions. Colonies were either randomly picked up or when the plate contained less than 10 colonies (Leisner et al., 1997). The purity of the isolates was checked by streaking again to fresh agar plates, followed by macroscopic and microscopic examinations. The strains displaying the general characteristics of lactic acid bacteria were chosen from each plate for further studies. The strains of lactic acid bacteria were stored without appreciable loss of properties in skimmed milk at -20°C. Working cultures were also kept on MRS agar or M17 agar slant at 4°C and streaked every 4 weeks (Samelis et al., 1994; Herrero et al., 1996).
Identification of lactic acid bacteria to the genus level: For identification of lactic acid bacteria, overnight cultures of each isolate in MRS broth (Oxoid) were used. All isolates were initially tested for Gram reaction, catalase enzyme and production of acid from glucose in Hugh and Leifson medium by oxidation or fermentation reaction (Harrigan and MacCance, 1976). Only Gram positive bacteria with catalase negative reactions were observed (Schillinger and Lucke, 1987; Garvie, 1986; Kandler and Weiss, 1986) and the representative isolates were purified by successive streaking onto the same agar substrate. For the Gram-positive, catalase negative rods, growth at various temperatures 10°, 15° and 45°C, tolerance of different salt levels (2, 4 and 6.5% w/v NaCl, hetero- and homo-fermentative activity (using MRS broth) with inverted Durham tubes in MRS broth were determined. Twenty isolates from raw cow milk were then selected based on the above tests for further identification. The isolates were stored at -40°C in MRS broth containing 10% glycerol. The bacteria were characterized by microscopic and by conventional biochemical and physiological tests. The cultures were examined for colony and cell morphology, motility, Gram stain and production of acid from glucose (Harrigan and MacCance, 1976), in addition to the oxidation and fermentation test according to Hugh and Leifson (1953). These preliminary tests make it possible to classify the isolates in genus on the basis of characteristic and tests of identification mentioned by Harrigan and MacCance (1976), Hammes et al. (1992), Holzapfel and Schillinger (1992) and Dicks et al. (1993).
Identification of lactic acid bacteria to the species level: After their microscopic examination, Gram +ve and catalase –ve lactobacilli were tested for their sugar fermentation pattern, production of ammonia from arginine in addition to their ability of growth at 15 and 45°C according to Harrigan and MacCance(1976). The carbohydrate fermentation profiles of the selected 20 isolates were investigated using API 20A and API CHL medium according to manufacture’s instructions (API System, Bio-Merieux, France). The bacteria identified by the use of a computer programme, API LAB PLUS, version 3.2.2 software (BioMerieux) and reference to Bergey’s Manual of Systematic Bacteriology.
RESULTS
Identification of isolates: The physiologic characteristics of the strains are shown in Table 1. Out of a total of 37 isolates obtained from raw cow milks from different areas of Khartoum state, twenty-three strains showed positive Gram reactions, absence of mobility, absence of spore formation,absence of catalase activity, cocci which produced no gas from glucose (12 isolates) and/or rods (11 isolates).
| Table 1: | Phenotypic characteristics of lactic acid bacteria isolated from cow's raw milks |
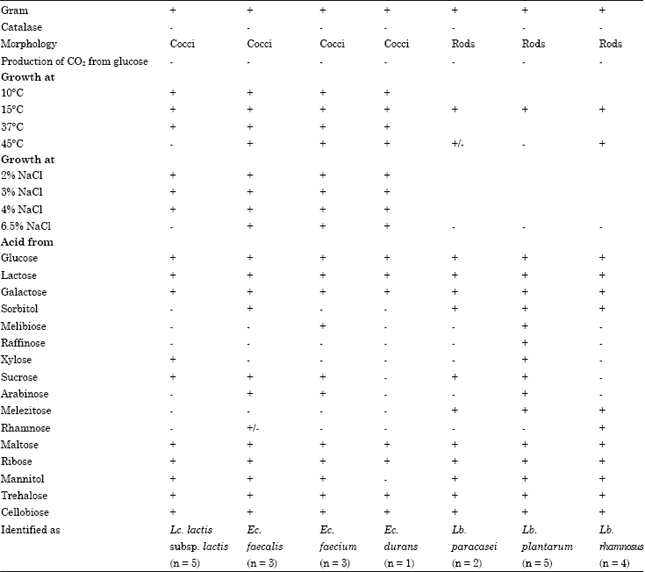 | |
| +: Positive reaction, -: Negative reaction, -/+: Variable reaction | |
Among the cocci, 5 isolates were able to grow at 10 and 37°C, but none at pH 9.6 broth and 45°C. In addition, they did not survive at 60°C for 30 min. All of the 5 strains could grow in 4%, but not in 6.5% NaCl broth and produced NH3 from arginine. The strains formed acid from lactose and ribose but acid production from mannitol, sucrose and xylose was strain dependent.
DISCUSSION
The phenotypic characteristics of the strains (Table 1) suggest their close resemblance to Lc. lactis subsp. lactis (Sharpe, 1979; Schleifer et al., 1985; Balows et al., 1991). Seven isolates of cocci were able to grow at 10 and 45°C in 6.5% NaCl and pH 9.6 broth. They also survive at 60°C for 30 min and form NH3 from arginine, but not CO2 from glucose and were characterized as enterococci. Three of them seemed to be Ec. faecalis, as suggested by their ability to survive at 60°C for 30 min and to ferment sorbitol (Sharpe, 1979; Schleifer and Kilpper-Balz, 1984; Devriese et al., 1993; Manero and Blanch, 1999). Four enterococci strains were differentiated by their ability to form acid from sugars. Thus, 3 strains producing acid from mannitol and Arabinose were characterized as Ec. faecium, one strain is characterized by inability to ferment melibiose and sucrose and unable, in general, to ferment sugars as Ec. durans (Schleifer and Kilpper-Balz, 1984; Devriese et al., 1993; Manero and Blanch, 1999). The 11 isolates of Gram-positive rods grew at 15°C and did not form either CO2 from glucose or NH3 from arginine. These characteristics suggest their classification as facultatively heterofermentative lactobacilli (Sharpe, 1979; Balows et al., 1991). Two out of 11 isolates did not form acid from arabinose, melibiose, raffinose and rhamnose and were characterized as Lb. paracasei subsp. pracasei (Collins et al., 1991; Balows et al., 1991). A total of 5 isolates of rods were classified as Lb. plantarum, as suggested by their sugar fermentations patterns. All these strains fermented arabinose, cellobiose, lactose, maltose, melibiose, raffinose, ribose, sucrose and trehalose (Sharpe, 1979; Balows et al., 1991). These strains did not form acid from rhamnose and acid production from sorbitol and xylose was variable and strain dependant. The last 4 strains were unable to ferment melibiose, raffinose, xylose, sucrose and arabinose but were able to form acid from rhamnose and classified as Lb. rhamnosus. Lactobacilli isolates represent a significant part among our isolates and are represented by Lb. plantarum, L. rhamnosus and Lb. paracasei. These species are frequently isolated from raw milk and dairy products (Tsakalidou et al., 1994). In other works, Mathara et al. (2004) and Abdelgadir et al. (2001) isolated Lactobacillus plantarum, Lactobacillus acidophilus and Lb. rhamnosus and Lb. fermentum from fermented products and showed that these species represent more than 60% of the isolated lactobacilli. In addition, Medina et al. (2001) showed that 8% of Lactobacillus isolated from ewe's milk and cheese in North from Argentina belonged to Lb. acidophilus.
Generally, the species identified in the present study, were in good agreement with other studies. Lactobacillus plantarum, Lactococcus lactis ssp. lactis, Lactobacillus delbrueckii subsp. lactis, Leuconostoc lactis and Leuconostoc citreum were identified in South African traditional fermented milks (Beukes et al., 2001). Lactococcus lactis ssp. lactis, Lactobacillus plantarum and Lactobacillus delbrueckii subsp. lactis were identified in Zimbabwean fermented milk (Feresu and Muzondo, 1990). Lactobacillus plantarum, Lactococcus lactis ssp. lactis and Lactobacillus confus were identified in Masaï fermented milk in Northern Tanzania (Isono et al., 1994).
CONCLUSION
The obtained results demonstrated that there is a diversity of lactic acid bacteria in cow's raw milk from cow in Khartoum state, Sudan. This local raw milk could serve as source for beneficial lactic acid bacteria in future researches.
REFERENCES
- Abdelgadir, W.S., S.H. Hamad, P.L. Moller and M. Jakobsen, 2001. Characterisation of the dominant microbiota of Sudanese fermented milk Rob. Int. Dairy J., 11: 63-70.
CrossRef - Aukrust, T. and H. Blom, 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int., 25: 253-261.
CrossRef - Beukes, E.M., B.H. Bester and J.F. Mostert, 2001. The microbiology of South African traditional fermented milks. Int. J. Food Microbiol., 63: 189-197.
CrossRefPubMedDirect Link - Caplice, E. and G.F. Fitzgerald, 1999. Food fermentations: Role of microorganisms in food production and preservation. Int. J. Food Microbiol., 50: 131-149.
CrossRefPubMedDirect Link - Carr, F.J., D. Chill and N. Maida, 2002. The lactic acid bacteria: A literature survey. Crit. Rev. Microbiol., 28: 281-370.
CrossRefDirect Link - Collins, M.D., U. Rodriquez, C. Ash, M. Aguirre and J.A.E. Farrow et al., 1991. Phylogenic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol. Lett., 77: 5-12.
Direct Link - De Man, J.C., M. Rogosa and M.E. Sharpe, 1960. A medium for the cultivation of Lactobacilli. J. Applied Bacteriol., 23: 130-135.
CrossRefDirect Link - Devriese, L.A., B. Pot and M.D. Collins, 1993. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically dinstinct enterococcal species and species groups. J. Applied Bacteriol., 75: 399-408.
PubMed - Dicks, L.M.T., L. Fantuzzi, F.C. Gonzales, M. du Toit and M. Dellaglio, 1993. Leuconostoc argentinum sp.nov isolated from argentine raw milk. Int. J. Syst. Bacteriol., 43: 347-351.
CrossRefDirect Link - Feresu, S. and M.I. Muzondo, 1990. Identification of some lactic acid bacteria from two Zimbabwean fermented milk products. World J. Microbiol. Biotechnol., 6: 178-186.
Direct Link - Gobbetti, M. and A. Corsetti, 1997. Lactobacillus sanfrancisco a key sourdough lactic acid bacterium: A review. Food Microbiol., 14: 175-188.
CrossRef - Hammes, W.P., N. Weiss and W.H. Holzapfel, 1992. The Genera Lactobacillus and Carnobacterium. In: The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications, Balows, A., H.G. Truper, M. Dworkin, W. Harder and K.H. Schleifer (Eds.). 2nd Edn., Vol. 2, Springer-Verlag, New York, USA., ISBN: 9783540972587, pp: 1534-1593.
- Harrigan, W.F. and M.E. McCance, 1976. Laboratory Methods in Food and Dairy Microbiology. 1st Edn., Academic Press, London, pp: 25-29.
Direct Link - Harris, L.J., H.P. Fleming and T.R. Klaenhammer, 1992. Characterization of two nisin producing Lactococcus lactis subsp. lactis strains isolated from a commercial sauerkraut fermentation. Appl. Environ. Microbiol., 58: 1477-1483.
Direct Link - Hugh, R. and E. Leifson, 1953. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram negative bacteria. J. Bacteriol., 66: 24-26.
CrossRefDirect Link - Isono, Y., I. Shingu and S. Shimizu, 1994. Identification and characteristics of lactic acid bacteria from masi fermented milk in Northern Tanzania. Biosc. Biotechnol. Biochem., 58: 660-664.
Direct Link - Leisner, J.J., G. Rusul, B.W. Wee, H.C. Boo and K. Mohammad, 1997. Microbiology of chili bo, a popular Malaysian food ingredient. J. Food Protect., 60: 1235-1240.
Direct Link - Lonvaud-Funel, A., 2001. Biogenic amines in wines: Role of lactic acid bacteria. FEMS Microbiol. Lett., 199: 9-13.
CrossRefDirect Link - Liu, S.Q., 2003. Review article: Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int. J. Food Microbiol., 83: 115-131.
CrossRef - Manero, A. and A.R. Blanch, 1999. Identification of Enterococcus ssp. with a biochemical key. Applied Environ. Microbiol., 65: 4425-4430.
Direct Link - Mathara, J.M., U. Schillinger, P.M. Kutima, S.K. Mbugua and W.H. Holzapfel, 2004. Isolation identification and characterization of the dominant microorganisms of kule naoto: The Maasai traditional fermented milk in Kenya. Int. J. Food Microbiol., 94: 269-278.
PubMedDirect Link - Medina, R., M. Katz, S. Gonzalez and G. Oliver, 2001. Characterization of the lactic acid bacteria in ewe`s milk and cheese from northwest Argentina. J. Food. Prot., 64: 559-563.
PubMedDirect Link - O'Sullivan, L., R.P. Ross and C. Hill, 2002. Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie, 84: 593-604.
CrossRefPubMedDirect Link - Samelis, J., F. Maurogenakis and J. Metaxopoulos, 1994. Characterization of lactic acid bacteria isolated from naturally fermented Greeck dry salami. Int. J. Food. Microbiol., 23: 179-196.
PubMedDirect Link - Schillinger, U. and F.K. Lucke, 1987. Identification of lactobacilli from meat and meat product. Food Microbiol., 4: 199-208.
CrossRef - Schleifer, K.H. and R. Kilpper-Balz, 1984. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int. J. Syst. Bacteriol., 34: 31-34.
CrossRef - Terzaghi, B.E. and W.E. Sandine, 1975. Improved medium for lactic streptococci and their bacteriophages. Applied Microbiol., 29: 807-813.
PubMedDirect Link - Tsakalidou, E., E. Manolopoulou, E. Kabaraki, B. Pot, K. Karel and G. Kalantzopoulos, 1994. The combined of whole-cell protein extracts for the identification (SDS-PAGE) and enzyme activity screening of lactic acid bacteria isolated from traditional dairy products. Syst. Applied Microbiol., 17: 444-449.
Direct Link








