ABSTRACT
This study aimed to detect the allelic frequency and identification of the genetic variants of α S1-casein gene in Egyptian goat using PCR-RFLP method. Forty-four animals used for detecting the presence of 10 polymorphisms of goat α S1-casein. The PCR products were digested by restriction enzyme XmnI and resolved using gel electrophoresis. Present results showed that Egyptian goat breeds carry low percentage of homozygous strong alleles A (4.5%) and B (6.8%) of milk casein used for cheese industry. The mild allele D recorded very low percentage 2.3%. This allele associated with medium level of milk protein favorable for allergic nutritional purposes. In conclusion, the allele frequencies and identification for animals α S1-casein are important for identifying animals carrying strong and mild alleles needed for improving a given trait depend on the frequency of favorable allele. The more will gained by selecting for strong and mild alleles of α S1-casein. So that, the breeding strategies should be changed toward milk protein alleles required for economic milk industry.
PDF Abstract XML References
How to cite this article
DOI: 10.3923/ijds.2006.27.31
URL: https://scialert.net/abstract/?doi=ijds.2006.27.31
INTRODUCTION
The fluid milk and other dairy products have been essential components of the human diet in a variety of cultures, mainly because the high quality of the protein contained in milk and the variety of minerals and vitamins. It is known that differences in the primary structure of milk proteins may effect, directly or indirectly, milk properties (Grosclaude, 1988). Little attention has been paid to the possibility of changing the manufacturing properties of milk through genetic selection, particularly when compared with what has been achieved in increased genetic potential for milk yield.
The relative amounts of the four casein fractions (α S1, β, αS2 and K) affect the physiochemical, nutritional and technological properties of milk of domestic ruminants (Ramunno et al., 2000). The first description of polymorphism in milk protein was reported for two variants β-lactoglobulin in cow’s milk (Aschaffenburg and Drewery, 1955). Since then, many other polymorphic have been characterized, both in caseins and milk whey protein, especially over the last decade, owing to the improvement of both biochemical and molecular techniques.
The molecular analysis of the goat genome is currently carried out using specific goat sequences as well as heterologous ones isolated from cattle and sheep (Vaiman et al., 1996). Deoxyribonucleic Acid (DNA) based markers have a number of favorable characteristics which represent ideal tool for studying the genetic quantitative traits (Ajmone-Marsan et al., 2001).
The goat α S1-casein (CSN1S1) locus is characterized by at least 13 alleles associated with four different levels synthesis: high, medium, low and null (Martin et al., 1999). The goat homozygous for alleles associated with a high content of α S1-casein produce milk characterized by a minor diameter of micelles, significantly higher percentage of protein, fat, total calcium and better parameters for curd firming time, curd firmness and cheese yield compared with goats homozygous for alleles associated with low or intermediate content (Martin et al., 1999; Remefu et al., 1989; Manfredi et al., 1993; Remeuf et al., 1993; Grosclaude et al., 1994; Pirisi et al., 1994; Remeuf et al., 1995; Lamberet et al., 1996).
This study aimed to detect the allelic frequency and identification of the genetic variants of α S1-casein gene in Egyptian goat using PCR-RFLP method.
MATERIALS AND METHODS
Genomic DNA Extraction
Genomic DNA was extracted from whole blood by phenol-chloroform method with minor modification (John et al., 1991). Ten milliliter of blood taken on EDTA was mixed with 25 mL cold sucrose-triton and the volume was completed to 50 mL by autoclaved double distilled water. The solution was mixed well and the nuclear pellet was obtained by spin and discarding the supernatant. The nuclear pellet was suspended in lysis buffer (10 mM Tris base, 400 mM NaCl and 2 mM sodium EDTA) pH 8.2, with 20% Sodium Dodecyl Sulfate (SDS) and proteinase K (10 mg mL-1) and incubated overnight in a shaking water-bath at 37°C.
Nucleic acids were extracted once with phenol, saturated with Tris-EDTA (TE) buffer (10 mM Tris, 10 mM NaCl and 1mM EDTA), followed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) until there was no protein at the interface. This was followed by extraction with chloroform-isoamyl alcohol (24:1).
To each extraction, equal volumes of the solvent were added, followed by thorough mixing and centrifugation for 10 min at 2000 rpm. The top layer was carefully transferred to another Falcon tube for the next extraction. To the final aqueous phase, 0.1 volume of 2.5 M Na acetate and 2.5 volume of cold 95% ethanol were added. The tubes were agitated gently to mix the liquids and a fluffy white ball of DNA was formed. The DNA was picked up with a heat-sealed Pasteur pipette and washed briefly in 70% ethanol.
The DNA was finally dissolved in an appropriate volume of 1X TE buffer. DNA concentrations were determined and diluted to the working concentration of 50 ng μL-1, which is suitable for polymerase chain reaction.
Polymerase Chain Reaction (PCR)
The primer used in this study was synthesized from published nucleotide sequence of goat α S1-casein gene (forward: 5´ TTCTAAAAGTCTCAGAGGCAG 3´), (reverse: 5´ GGGTTGATAGCCTTGTATGT 3´) (Ramunno et al., 2000). Its PCR product enable us to detect 10 genetic variants of αS1-casein after digestion with a specific restriction enzyme.
A PCR cocktail consists of 1.0 (μM upper and lower primers and 0.2 mM dNTPs, 10 mM Tris (pH 9), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin (w/v), 0.1% Triton X-100 and 1.25 units of Taq polymerase. The cocktail was aliquot into tubes with 100 ng DNA of goat. The reaction mixture was overlaid with sterile mineral oil and is run in a Coy Temp Cycler II (Coy Corporation, MI, USA) the amplification protocol consisted of 30 cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 2 min.
RFLP and Agarose Gel Electrophoresis
Twenty microliter of PCR product were digested with 10 units of restriction enzyme XmnI used in a final reaction volume 25. The reaction mixture was incubated at 37°C in water bath over night. After restriction digestion, the restricted fragments were analyzed by electrophoresis on 2.5% agarose/1X TBE gel stained with ethidium bromide. The 100 bp ladder was used as molecular size marker. The bands were visualized under UV light and photographed with yellow filter on black and white film.
Results and Discussion
Forty four animals used for detecting the presence of 10 polymorphisms of goat αS1-casein. The PCR products were digested by restriction enzyme XmnI and resolved using gel electrophoresis. The restriction fragment length polymorphism determined by XmnI allowed to identify the genotypes a/a at 150 bp, b/b at 161 bp, c/c at 212 bp, d/d at 223 bp.
| Table 1: | Genotype frequencies of α S1-casein after XmnI digestion of fragment obtained by PCR products of Egyptian goat DNA |
 | |
| Table 2: | Genotype frequencies of α S1-casein among different Egyptian goat breeds |
 | |
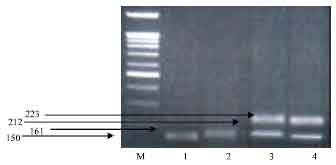 | |
| Fig. 1: | DNA electrophoretic pattern obtained after digestion of PCR amplified of goat casein α 1 (CSN1S1) products with XmnI, M: 100-bp ladder marker, Lane 1: homozygous a/a genotype, Lane 2: homozygous b/b genotype, Lane 3: heterozygous b/d genotype, Lane 4: heterozygous a/c genotype |
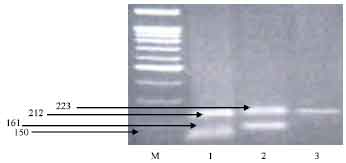 | |
| Fig. 2: | DNA electrophoretic pattern obtained after digestion of PCR amplified of goat casein α 1 (CSN1S1) products with XmnI, M: 100-bp ladder marker, Lane 1: heterozygous a/c genotype, Lane 2: heterozygous b/d genotype, Lane 3: homozygous d/d genotype |
The presence of a/b should be illustrated by two fragments at 150 and 161 bp, a/c by two fragments at 150 and 212 bp, a/d by two fragments at 150 and 223 bp, b/c by two fragments at 161 and 212 bp, b/d by two fragments at 161 and 223 bp, c/d by two fragments at 212 and 223 bp (Table 1).
Among different breeds (Table 2), the results of genetic variants showed that the Damascus breed has five different genotypes a/a, b/b, d/d, a/c, b/d (Fig. 1 and 2), the Sahrawi breed has a/c with high frequency 90.9%, while the Baladi breed carries two different genotypes with different frequencies (Table 2). In Zaraibi breed, the genotype b/d is the most common by 80%.
In caprine breeds, four types of alpha S1-casein alleles are identified and associated with various amounts of protein in milk. The A, B and C are strong alleles associated with a high level of αS1-casein in milk (3.5 g L-1). The D and E alleles with a medium level (1.1 g L-1), the F and G alleles with a low level (0.45 g L-1). The contribution of null allele is very low or non existent of α S1-casein (Ramunno et al., 2000). The quantity of total caseins in caprine milk is positively correlated with the amount of α S1-casein. Therefore, milk from animals possessing strong alleles contain significantly more total caseins than milk from animals without those alleles. This is important because animals with mild alleles can be employed to produce milk for allergic subjects, while the other animals can be used to produce milk for the dairy industry (Martin et al., 1999; Leroux et al., 1990; Roncada et al., 2002).
Present results revealed that Egyptian goat breeds showed low percentage of homozygous strong alleles A (4.5%) and B (6.8%) of milk casein used for cheese industry. The mild allele D recorded very low percentage 2.3%. This allele associated with medium level 1.1 g L-1 of milk protein favorable for allergic subject (nutritional purposes) especially the infant diet where the goat milk with low casein is reported less allergenic than cow’s milk (Roncada et al., 2002). These results indicated that alleles require for economic milk industry present with low percentage in Egyptian goat breeds.
In conclusion, the alleles frequencies and identification for animals α S1-casein are important for identifying animals carrying the strong and mild alleles needed for improving a given trait depend on the frequency of favorable allele. The more will gained by selecting for strong and mild alleles of α S1-casein. So that, the breeding strategies should be changed toward milk protein alleles required for economic milk industry.
ACKNOWLEDGMENTS
Especial thanks to Prof. Dr. Soheir M. El Nahas and Dr. Othman E. Othman for their support. This research was conducted as a part of the project Genetic polymorphism of some genes related to quantitative traits in different breeds of goat in Egypt (2002-2005), which was supported from the National Research Center (Egypt).
REFERENCES
- Ajmone-Marsan, P., R. Negrini, P. Crepaldi, E. Milanesi, C. Gorni, A. Valentini and M. Cicogna, 2001. Assessing genetic diversity in Italian goat populations using AFLP® markers. Anim. Gen., 32: 281-288.
CrossRefDirect Link - John, S.W.M., G. Weitzner, R. Rozen and C.R. Scriver, 1991. A rapid procedure for extracting genomic dna from leukocytes. Nucleic Acid Res., 19: 408-412.
Direct Link - Lamberet, G., C. Degas, A. Delacroix-Buchet and L. Vassal, 1996. Effect of characters linked to A and F caprine α S1-casein alleles on goat flavours: Cheese making with protein-fat exchange. Lait, 76: 349-361.
Direct Link - Leroux, C., P. Martin, M. Mahe, H. Leveziel and J. Mercier, 1990. Restriction fragment length polymorphism identification of goat alpha S1-casein alleles: A potential tool in selection of individuals carrying alleles associated with a high level protein synthesis. Anim. Gen., 21: 341-351.
Direct Link - Martin, P., M. Ollivier-Bousquet and F. Grosclaude, 1999. Genetic polymorphism of caseins: A tool to investigate casein micelle organization. Int. Dairy J., 9: 163-171.
Direct Link - Pirisi, A., O. Colin, F. Laurent, J. Scher and M. Parmentier, 1994. Comparison of milk composition, cheesemaking properties and textural characteristics of the cheese from two groups of goats with a high or low rate of αS1-casein synthesis. Int. Dairy J., 4: 329-345.
CrossRefDirect Link - Roncada, P., A. Gaviraghi, S. Liberatori, B. Canas, L. Bini and G. Greppi, 2002. Identification of caseins in goat milk. Proteomics, 2: 723-726.
CrossRefPubMedDirect Link - Vaiman, D., L. Schibler, F. Bourgeois, A. Oustry, Y. Amigues and E.P. Cribiu, 1996. A genetic linkage map of the male goat genome. Genetics, 144: 279-305.
Direct Link








