ABSTRACT
DNA methylation is one of three epigenetic mechanisms which has seen increasing interest among cancer researchers, as changes in DNA methylation have emerged as one of the most consistent molecular aberrations in various neoplasms. Aberration of DNA methylation have been identified in the global genomic DNA (hyper or hypo methylation) or within CpG islands in promoter regions of tumor suppressor genes. Several types of bacteria which cause chronic infections are associated with cancer development and chronic infection of Helicobacter pylori might give rise to changes in pattern of DNA methylation. Recent discoveries are giving evidence in the involvement of aberration in DNA methylation in conferring resistance to antitumor agents, and efforts of molecular biologists are directed to overcome this problem by introducing new epigenomic drugs which reform DNA methylation, and help in tumor therapy. Another interesting prospective of current research in DNA methylation is the possibility of development molecular markers for early detection of tumors. The objectives of this article are to review various recent progress of DNA methylation in cancer research.
PDF Abstract XML References Citation
How to cite this article
DOI: 10.3923/ijcr.2010.188.201
URL: https://scialert.net/abstract/?doi=ijcr.2010.188.201
INTRODUCTION
The term epigenetics was introduced in 1940s by Waddington to show the way genes and their products bring the phenotype into being, nowadays the term is used to clarify the mechanisms through which cells become dedicated and committed to a particular form, structure, phenotype or function and to explain how genetic changes in gene expression whether activation or inactivation, are caused by mechanisms other than changes in the DNA sequence (Holliday, 1990; Jablonka and Lamb, 2002). Scientists consider the epigenetic inactivation of genes as an important a driving force of gene malfunction as the inactivation of genes by mutation (Esteller and Herman, 2002).
Currently, scientific community engaged in molecular genomic research coined other related term epigenomics from epigenetics and genome, which deals with molecular elements and mechanisms influencing gene expression. This new discipline gives new potentials and novel insights into the genomic research, because of its capability to detect quantitative alterations, multiplex modifications and regulatory sequences outside of genes (Callinan and Feinberg, 2006). Nowadays epigenomics is at the epicenter of modern molecular medical biotechnology and it can help to explain the relationship between an individual's genetic background, environment, development, aging and disease. It can do so because the epigenetic state varies among tissues, during lifetime and progression of disease, whereas the DNA sequence remains essentially the same (Heindel et al., 2006). Moreover, unlike genomic DNA sequence, epigenomic programs are more susceptible to environmental effects on the cells; these can modify the functions of genes leading to deleterious phenotypic expression (Guil and Esteller, 2009).
Molecular biologists have identified three main epigenomic mechanisms which are involved in the gene expression that are not caused by DNA sequence alterations. These mechanisms are DNA methylation, histone modifications and RNA interference (Shilatifard, 2006; Suzuki and Yoshino, 2008; Waterland and Michels, 2007). Taking into account DNA methylation is one of unique epigenomic mechanisms associated with neoplasia and the rapid progress in research work in this area, the focus of this study is to review the perspective of DNA methylation in cancer research.
Discovery of DNA Methylation
DNA methylation is the addition of a methyl group to the carbon-5 position of cytosine residues and is considered the only common covalent modification of human DNA and occurs almost exclusively at cytosine that is followed immediately by a guanine, so-called CpG dinucleotide (Strathdee and Brown, 2002). However, there are other two types of methylated bases are known to occur: N6-methyladenine, which is found in bacteria and eukaryotes (Dunn and Smith, 1958) and N4-methylcytosine, a minor component of bacterial DNA (Ehrlich et al., 1987). But C5-methylcytosine is the most prevalent of the known DNA modifications in the genomic DNA of mammals and plants and is considered the fifth nucleotide (Adams, 1990); there are indications in the literature that fifth base was first described in DNA isolated from the tubercle bacillus (Johnson and Coghill, 1925) and in calf thymus DNA (Hotchkiss, 1948).
In human beings, the main epigenetic modification is methylation of the cytosine and thus formation of 5-methylcytosine. Published results showed 3-4% of all cytosine molecules are methylated and the resulting 5-methylcytosine makes up 0.75-1% of all nucleotide bases in the DNA of normal human tissue (Esteller et al., 2001; Fraga et al., 2002). Stretches of CpG dinucleotides are concentrated in regions called CpG islands, which are generally unmethylated. These motifs span the 5' end of many genes including the promoter, untranslated region and exon 1, which makes them excellent indicators of where a gene begins (Bird, 1986).
Addition of the methyl group at cytosine ring of 5¯-CpG-3¯ sequence is catalyzed by one of three DNA methyl transferases (Dnmt1, Dnmt3a and Dnmt3b) with S-adenosyl methionine as the methyl donor (Bird, 1992). The Dnmt3 family establishes the initial CpG methylation pattern de novo, whereas Dnmt1 maintains this pattern during chromosome replication (Chen and Li, 2006; Cheng and Blumenthal, 2008; Hermann et al., 2004).
Functions of DNA Methylation
DNA methylation in prokaryotes is not an essential system for viability since nul mutants in bacteria are viable; however bacteria had established possible regulatory function of DNA methylation and it was possible to isolate an enzyme from E. coli which could methylate unmodified DNA at specific sites and confer resistance to the homologous restriction endonuclease (Kuehnlein and Arber, 1972; Jost and Saluz, 1993).
Recognition of C5-methylcytosine possible physiological role in eukaryotes was first suggested in 1964 (Srinivasan and Borek, 1964). Since then several functions were reported for DNA methylation in human genome, it is essential for the maintenance of X chromosome inactivation and imprinting (Tucker et al., 1996). Outside of these two unique situations, however, the role of CpG methylation (which affects up to 70% of CpG sites) remains under extensive investigation by researchers. It has been noted that most CpG sites in the human genome are actually present within parasitic DNA sequences (sequences that have high rates of recombination), including Alu and Line elements and it was proposed that DNA methylation evolved as a defense mechanism against harmful consequences of these sequences, such as recombination events (Baccarelli et al., 2010; Yang et al., 2004). This suggestion was reinforced and supported by demonstrating that most Alu sequences are greatly methylated in the human genome and that such methylation inhibits Alu transcription (Kidwell and Lisch, 2001). Interestingly, other data suggested that Alu transcription could be activated by cellular stress and that Alu RNA might inhibit apoptosis through inhibition of the double-stranded RNA-activated kinase PKR (Schmid, 1991, 1998).
Several studies suggested some beneficial effects of DNA methylation, it had been proposed that Alu demethylation in cancer might lead to a survival advantage through inhibited apoptosis (Schmid, 1998), while DNA methylation in the Epstein-Barr Virus (EBV) genome contributes to maintaining latency of the virus and that EBV might use this methylation to prevent the expression of viral antigens and thus escape immune surveillance (Elliott et al., 2004). However, malfunction in DNA methylation mechanisms can lead to various deleterious outcomes in development, aging and diseases and direct association between DNA methylation aberration and cancer has been reported.
DNA Methylation and Cancer
In the past three decades, there has been an enormous amount of research work on the possible association of DNA methylation with various malagnancies. Thus, DNA methylation has seen a stream of interest among cancer researchers as it has become known as one of the most consistent molecular aberrations in neoplasms. Molecular studies have shown strong evidence which has correlated changes in normal pattern of DNA methylation with various types of cancers. It is increasingly clear and accepted nowadays, that aberrant DNA methylation is the most common molecular lesion of cancer cell, for example, global hypomethylation leads to oncogene activation and chromosomal rearrangement; in addition, hypermethylation is observed in genomes of various types of cancers (Liu et al., 2003; Nguyen et al., 2001).
Leukaemia has traditionally been viewed as a genetic disease; however other studies showed DNA methylation defects also play an important role. A recent study illustrated that digestion of genomic DNA of normal individuals with HpaII (which cleaves the sequence CCGG only if the internal cytosine residue is unmethylated) and MspI (which cleaves the same sequence regardless of methylation) showed lower degree of differences in RAPD-DNA patterns, indicating normal methylation patterns. However, different picture was shown when studying RAPD-DNA bands profile of CML genomic DNA following digestion with restriction HpaII and MspI (Fig. 1), clear variations in bands patterns were observed when using selected RAPD primers to amplify digested genomic DNA by two enzymes (MspI and HpaII) for normal individuals and CML patients (Ibrahim et al., 2010a).
An interesting investigation was carried out on the breast cancer cell lines by Roll et al. (2008) to investigate the relationship between promoter methylation (assessed by methylation-specific PCR, bisulfite sequencing and 5-aza-2'deoxycytidine treatment) and the DNA methyltransferase machinery (total DNMT activity and expression of DNMT1, DNMT3a and DNMT3b proteins). The study revealed two groups of cell lines that possessed distinct methylation signatures:
| • | Hypermethylator cell lines |
| • | Low-frequency methylator cell lines |
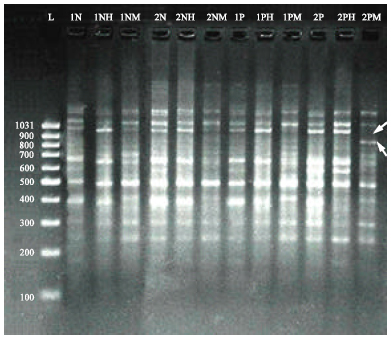 | |
| Fig. 1: | RAPD-PCR patterns of genomic DNA normal and CML patients obtained with decamer RAPD primers (OPW-17) before and after digestion with HpaII and MspI. Electrophoresis was performed on (1.5%) agarose gel and run with 3 volt cm-1 for 2 h. 1N and 2N: Amplified products of undigested genomic DNA extracted from normal individuals, 1P and 2P: Amplified products of undigested genomic DNA extracted from CML patients, H: Amplified products of digested genomic DNA with HpaII, M: Amplified products of digested genomic DNA with MspI, L: Lambda DNA (100-1031 bp). Arrows indicate addition and missing DNA fragments |
The hypermethylator cell lines were characterized by high rates of concurrent methylation of six genes (CDH1, CEACAM6, CST6, ESR1, LCN2, SCNN1A), whereas the low-frequency methylator cell lines did not methylate these genes. Hypermethylator cell lines coordinately over express total DNMT activity and DNMT3b protein levels compared to normal breast epithelial cells. In contrast, most low-frequency methylator cell lines possess DNMT activity and protein levels that are indistinguishable from normal (Roll et al., 2008).
Bisulphite sequencing of genomic DNA methylation revealed, in the majority colon rectal tumors, that the methylation was unequally distributed within the MAL promoter; on the other hand MSP analysis of a region close to the transcription start point was shown to be hypermethylated; in contrast, only a minority of the normal mucosa samples displayed hypermethylation (Lind et al., 2008). Other investigators used RAPD-PCR analysis to study aberration of DNA methylation of colon cancers; Luo et al. (2003) were able to get reproducible RAPD-PCR fingerprints from colon cancers patients generated by PCR amplification of genomic DNA with each RAPD primer or random primer pair, their results showed multiple changes that occurred in both the ACF (aberrant crypt foci) and tumor and additional alterations (gain of bands) that occurred only in the tumor when the RAPD bands are compared with those from normal crypts (Luo et al., 2003).
A study carried by Pfeifer and Rauch (2009) showed different pattern of DNA methylation in Lung cancer. In their study a genome-scale mapping technique for CpG methylation (MIRA-chip) was used, it was possible to characterize CpG island methylation and methylation patterns of entire chromosome arms at a level of resolution of 100 bp.
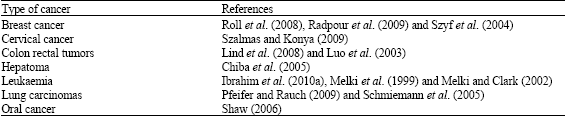
| Table 1: | Aberration of DNA methylation detected in various types of cancers |
 | |
In individual stage I lung carcinomas, several hundred and probably up to a thousand CpG islands become methylated. Interestingly, a large fraction (almost 80%) of the tumor-specifically methylated sequences is target of the Polycomb complex in embryonic stem cells. Homeobox genes are particularly overrepresented and all four HOX gene loci on chromosomes 2, 7, 12 and 17 are hotspots for tumor-associated methylation because of the presence of multiple methylated CpG islands within these loci. DNA hypomethylation at CpGs in squamous cell tumors preferentially affects repetitive sequence classes including SINEs, LINEs, subtelomeric repeats and segmental duplications. The investigators emphasized that since these epigenetic changes are found in early stage tumors, their contribution to tumor etiology as well as their potential usefulness as diagnostic or prognostic biomarkers of the disease should be considered.
The information shown in Table 1 gives a summary of the research work on the association of changes in DNA methylation in various types of cancers.
Methylation of Tumor Suppressor Genes
Tumor suppressor genes are genes that reduce the probability that human cell will turn into tumor cell and are known to play a critical role in regulating physiological processes when cells are allowed to divide and increase in number (Sherr, 2004; Yeo, 1999). When DNA damage is detected in a cell, some tumor suppressor genes can stop the cell from multiplying until the damage is repaired; however, once tumor suppressor genes do not function correctly, the cells with DNA damage continue to divide and can accumulate further DNA damage that can eventually lead to the formation of a cancer cell (Hussain et al., 2001; Macleod, 2000).
The first tumor-suppressor gene discovered was the Rb gene which is associated with retinoblastoma, a serious cancer of the retina that occurs in early childhood. In addition to retinoblastomas, mutations in the Rb gene have been detected in osteosarcomas, bladder carcinomas, small-cell lung carcinomas, prostate carcinomas, breast carcinomas, some types of leukemias and cervical carcinomas (Krug et al., 2002). Another tumor suppressor gene is p53, a gene found to be mutated in a large proportion of human cancers such as, in lung, colon, esophageal, ovarian, pancreatic, skin, stomach, head and neck, bladder, sarcoma, prostate, epatocellular, brain, breast, renal, thyroid, hematological malignancies, melanoma and cervical cancers; an increased amounts of cellular p53 protein after DNA damage have been associated with cell-cycle arrest and programmed cell death (apoptosis) and mutations or losses of p53 have been result in development of cancer (Velculescu and El-deiry, 1996; Foulkes, 2007). Hypermethylation within the promoter-associated CpG islands of suppressor genes could be particularly important, especially when this methylation affects tumor suppressor genes. Evidence for this came from the study of two such genes, VHL (the gene mutated in Von Hippel-Lindau disease) and CDKN2A (which encodes p16INK4A, a gene commonly mutated in many types of cancer) where, in a subset of tumors, dense promoter methylation was described, in association with (1) loss of gene expression (which could be partially restored using methylation inhibitors), (2) absence of coding region mutations and (3) tumor-specific patterns of methylation (Kuerbitz et al., 1999).
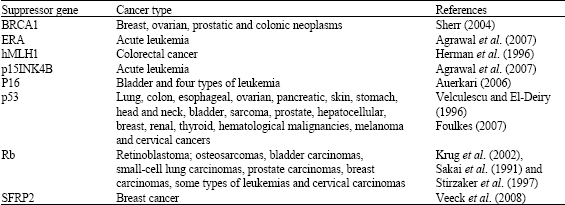
| Table 2: | Some tumor suppressor genes affected by DNA methylation in promoter region |
 | |
Others showed additional examples of this process of promoter methylation (including the hMLH1 gene, which is commonly mutated in colorectal cancer) and proposed that such hypermethylation should now be considered as equivalent to mutations and deletions in Knudson's two-step model of inactivation of tumor-suppressor genes (Herman et al., 1996).
Table 2 presents some of the known reported tumor suppressor genes which their expression are affected by DNA methylation of promoter regions.
DNA Methylation in Antitumor Drug Resistance
Chemotherapy resistance, either innate or acquired, requires expression changes in a large number of genes for its development; thus, it has been hypothesized that epigenetic-mediated changes could be the driving force responsible for chemotherapy resistance (Glasspool et al., 2006). Research group led by Candelaria had suggested that epigenetic changes are not solely a consequence of cancer cell-resistant phenotype, but that indeed these contribute to its development (Candelaria et al., 2007). It had been demonstrated that chemotherapeutic drugs induced DNA hypermethylation in cultured cells (Nyce et al., 1986; Nyce, 1989). Likewise, MCF-7 cells develop DNA hypermethylation when they acquire the multidrug-resistant phenotype and both hypermethylation and doxorubicin resistance are reversed by hydralazine or by antisense treatment against DNA methyltransferases (Segura-Pacheco et al., 2006). Kastl et al. (2010) were able to demonstrate that changes in the DNA methylation machinery are associated with resistance to docetaxel in breast cancer cells. Resistance to cisplatin is also accompanied by over-expression of DNA methyltransferases (Wang et al., 2001) and the resistance itself is induced by over-expression of DNA methyltransferase genes (Qiu et al., 2005). Recent study by Chang et al. (2010) provided further evidence that epigenetic promoter methylation is a frequent event during chronic cisplatin exposure and that secondary cahages in gene regulation can play an important role in generating drug resistant phenotypes, they suggested that differentially methylated genes, including those identified in their study (SAT, C8orf4, LAMB3, TUBB, GOS2 and MCAM) may provide informative drug resistance markers as well as therapeutic targets, potentially leading to improved therapies for cancer patients with better and more durable clinical responses (Chang et al., 2010).
The use of epigenetic therapies, as a strategy to overcome drug resistance, needs to be investigated more fully to determine their effectiveness in different cancers and for different chemotherapy drugs, hence, it could be expected that agents targeting DNA methylation, would by reverting the epigenetic marker and overcome chemotherapy resistance (Perez-Plasencia and Duenas-Gonzalez, 2006).
DNA Methylation Inhibitors in the Treatment of Cancer
In view of the growing interest epigenetic therapies, DNA methylation inhibitors have found applications in treatments of cancers and few other diseases. In the following the perspectives of two epigenomic antimumor drugs of azanucleosides family will be reviewed. First drug is 2-deoxy-5-azacytidine (decitabine), a cytosine nucleoside analog, was synthesized in 1964 and is considered DNA precursor, it was reported that the drug once incorporated into DNA, inhibits further DNA methylation (De Vos and Van Overveld, 2005). As a consequence, aberrantly silenced genes, including tumor suppressor genes, can be reactivated and expressed. Decitabine has demonstrated activity in a broad range of hematologic disorders, including: Acute Myelogenous Leukemia (AML), Chronic Myelogenous Leukemia (CML) (Kantarjian et al., 2003, 2006) and sickle cell anemia (Saunthararajah et al., 2003). Results of two studies suggested that repeated courses of low-dose decitabine induces cytogenetic remissions in a substantial number of patients with MDS and pre-existing chromosomal abnormalities; response being associated with improved survival compared with patients in whom the cytogenetically abnormal clone persists. Patients with high-risk chromosomal abnormalities may particularly benefit from this treatment (Christman, 2002; Mesa et al., 2009).
Second drug is 5-azacytosine (azacitidine), also an inhibitor of DNA methylation, the drug is a ribonucleic acid (RNA) precursor; chemically azacitidine differs from decitabine in structure only slightly by having a hydroxyl group that is lacking in decitabine; it is the first drug to be approved by the FDA in May 2004 for treating rare family of bone-marrow disorders and has been given orphan-drug status, a group of pharmaceutical agents which has been developed specifically to treat rare medical conditions known as orphan diseases (Kaminskas et al., 2005a, b). It is also a pioneering example of an agent that targets epigenetic gene silencing, a mechanism that is exploited by cancer cells to inhibit the expression of genes that counteract the malignant phenotype (Issa et al., 2005; Kaminskas et al., 2005a, b; Komashko and Farnham, 2010; Kantarjian et al., 2006; Kornblith et al., 2002).
The molecular mechanisms for incorporation of two drugs into DNA had been investigated. Azacitidine is phosphorylated by the enzyme uridine-cytidine kinase and is then incorporated into RNA. It is incorporated into DNA by conversion to a deoxyribose form by the enzyme ribonucleotide reductase, which converts ribose to deoxyribose. Decitabine is phosphorylated by deoxycytidine kinase and then incorporated into DNA. Both drugs are pro-drugs to 5-azadeoxycytidine triphosphate; however, their biochemical differences may allow one to work in a patient when the other does not (Christman, 2002; Mesa et al., 2009).
It is worth mentioned other studies described clinical trials with these agents in myelodysplasia and lung cancer and reported on a few responses that justify further research in this field (Santini et al., 2001), however recent reports give promising results for the epigenomic drugs in cancer therapy (Komashko and Farnham, 2010; Peedicayil, 2008).
Bacterial Infections, Cancers and DNA Methylation
Infections involving viruses, bacteria and schistosomes have been linked to higher risks of malignancy. As early as 1772, Mycobacterium tuberculosis was thought to cause malignancy (Parsonnet, 1995). It had been reported a substantial number of bacterial pathogens have been linked to cancer, it is estimated that over 15% of malignancies worldwide can be attributed to infections (Pisani et al., 1997).
| Table 3: | Bacterial infections associated with various types of cancer |
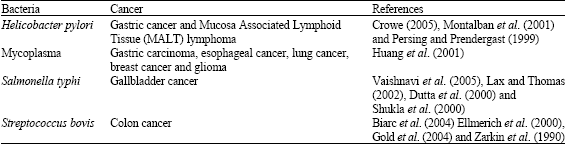 | |
The data presented in Table 3 showed various types bacteria associated with cancer. The most specific example of the inflammatory mechanism of carcinogenesis is Helicobacter pylori infection (Crowe, 2005; Parsonnet, 1995). Other studies reported high correlation between mycoplasma infection and different cancers, which suggested the possibility of an association between the two (Huang et al., 2001). Recent studies suggested bacteria have been linked to cancer by two mechanisms: induction of chronic inflammation and production of carcinogenic bacterial metabolites (Vakevainen et al., 2002), thus it was noted the effect of bacterial infections on promotion of cancer and DNA hypermethylation (Bobetsis et al., 2007; Salaspuro, 2003) and aberration of DNA methylation (Shin et al., 2010). These results support the correlation between aberration of DNA methylation caused by bacterial infections and cancer.
Methylated Genes as New Cancer Biomarkers
Various studies have suggested that measurement of the methylation status of the promoter regions of specific genes can aid early detection of cancer, determine prognosis and predict therapy responses (Zhu and Yao, 2009).
An interesting investigation led by Sanchez-Cespedes showed that aberrant DNA methylation at four genes in primary tumors from head and neck cancer patients might be utilized for identification of biomarkers and then possible use of the presence of this methylation as a marker for cancer cell detection in serum DNA. These four genes were tested by methylation-specific PCR and included: p16 (CDKN2A), O6-methylguanine-DNA-methyltransferase, glutathione S-transferase P1 and deathassociateprotein kinase (DAP-kinase). Fifty-five percent (52 of 95) of the primary tumors displayed promoter hypermethylation in at least one of the genes studied: 27% (26/95) at p16, 33% (31 of 95) at O6-methylguanine-DNA-methyltransferase; and 18% (17 of 92) at DAP-kinase. No promoter hypermethylation was observed at the glutathione S-transferase P1 gene promoter. The study detected a statistically significant correlation between the presence of DAP-kinase gene promoter hypermethylation and lymph node involvement and advanced disease stage. In 50 patients with paired serum available for epigenetic analysis, the same methylation pattern was detected in the corresponding serum DNA of 21(42%) cases. Among the patients with methylated serum DNA, 5 developed distant metastasis compared with the occurrence of metastasis in only 1 patient negative for serum promoter hypermethylation. The researchers concluded, promoter hypermethylation of key genes in critical pathways is common in head and neck cancer and represents a promising serum marker for monitoring affected patients (Sanchez-Cespedes et al., 2000).
Other possible DNA methylation biomarkers include the use of methylated GSTP1 for aiding the early diagnosis of prostate cancer, methylated PITX2 for predicting outcome in lymph node-negative breast cancer patients and methylated MGMT in predicting benefit from alkylating agents in patients with glioblastomas (Duffy et al., 2009). However, prior to clinical utilization, these findings require validation in prospective clinical studies.
Promoter hypermethylation of MAL gene as was shown as an early epigenetic diagnostic marker for colorectal tumors, hypermethylation was present in the vast majority of benign and malignant colorectal tumors and only rarely in normal mucosa, which makes it suitable as a diagnostic marker for early colorectal tumorigenesis (Lind et al., 2008).
It is worth mentioned, in this context, the recent results obtained by Ibrahim et al. (2009, 2010a, b) and Saleh et al. (2010) on possible use of specific DNA fragments detected by RAPD primers for detection of certain types of leukemia, their results showed that specific amplified DNA fragments could be identified in genome of cancer patients but are absent in normal genome.
CONCLUSION
Epigenomic mechanisms have important function in gene expression and any change has an impact on various biological phenomena, for example development, aging and diseases. DNA methylation is one of three important epigenomic mechanisms associated with various types of cancers. Genomic methylation changes associated with oncogenic transformation are detected and studying molecular basis of DNA methylation is considered vital and decisive in understanding how DNA methylation may contribute to tumorigenesis. Silencing of tumor suppressor genes is usually associated with hypermethylation in the promoter regions of cells, which results in loss of expression and subsequent tumorigenesis. Other factors have been reported to cause aberration of DNA methylation in the genomic DNA of cancer patients; these include chronic bacterial infections and resistance to antitumor drugs. Two types of DNA methylation inhibitors, azacitidine and decitabine, have generated much interest in cancer therapies.
ACKNOWLEDGMENT
The author would like to thank International Institute of Education (IIE) for financial support.
REFERENCES
- Adams, R.L., 1990. DNA methylation-the effect of minor bases on DNA protein interactions. Biochem. J., 265: 309-320.
PubMedDirect Link - Agrawal, S., M. Unterberg, S. Koschmieder, U. Stadt and U. Brunnberg et al., 2007. DNA methylation of tumor suppressor genes in clinical remission predicts the relapse risk in acute myeloid leukemia. Cancer Res., 67: 1370-1377.
Direct Link - Auerkari, E.I., 2006. Methylation of tumor suppressor genes p16(INK4a), p27(Kip1) and E-cadherin in carcinogenesis. Oral Oncol., 42: 4-12.
Direct Link - Baccarelli, A., L. Tarantini, R.O. Wright, V. Bollati and A.A. Litonjua et al., 2010. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics, 5: 222-228.
Direct Link - Biarc, J., I.S. Nguyen, A. Pini, F. Gosse and S. Richert et al., 2004. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S. bovis). Carcinogenesis, 25: 1477-1484.
Direct Link - Bobetsis, Y.A., S.P. Barros, D.M. Lin, J.R. Weidman and D.C. Dolinoy et al., 2007. Bacterial infection promotes DNA hypermethylation. Dent. Res., 86: 169-174.
PubMed - Callinan, P.A. and A.P. Feinberg, 2006. The emerging science of epigenomics. Hum. Mol. Genet., 15: R95-R101.
Direct Link - Candelaria, M., D. Gallardo-Rincon, C. Arce, L. Cetina and J.L. Aguilar-Ponce et al., 2007. A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann. Oncol., 18: 1529-1538.
Direct Link - Chang, X., C.L. Monitto, S. Demokan, M.S. Kim and S.S. Chang et al., 2010. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res., 70: 2870-2879.
Direct Link - Chen, T. and E. Li, 2006. Establishment and maintenance of DNA methylation patterns in mammals. Curr. Top. Microbiol. Immunol., 301: 179-201.
PubMed - Cheng, X. and R.M. Blumenthal, 2008. Mammalian DNA methyltransferases: A structural perspective. Structure, 16: 341-350.
PubMed - Christman, J.K., 2002. 5-Azacytidine and 5-aza-2-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene, 21: 5483-5495.
Direct Link - Chiba, T., O. Yokosuka, K. Fukai, Y. Hirasawa and M. Tada et al., 2005. Identification and investigation of methylated genes in hepatoma. Eur. J. Cancer, 41: 1185-1194.
PubMed - Crowe, S.E., 2005. Helicobacter infection, chronic inflammation and the development of malignancy. Curr. Opin. Gastroenterol., 21: 32-38.
Direct Link - Duffy, M.J., R. Napieralskic, J.W.M. Martensd, P.N. Spane and F. Spyratosf et al., 2009. Methylated genes as new cancer biomarkers. Eur. J. Cancer, 45: 335-346.
Direct Link - Dunn, D.B. and J.D. Smith, 1958. The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem. J., 68: 627-636.
Direct Link - Dutta, U., P.K. Garg, R. Kumar and R.K. Tandon, 2000. Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gallbladder. Am. J. Gastroenterol., 95: 784-787.
PubMed - Ehrlich, M., G.G. Wilson, K.C. Kuo and C.W. Gehrke, 1987. N4-methylcytosine as a minor base in bacterial DNA. J. Bacteriol., 169: 939-943.
Direct Link - Elliott, J., E.B. Goodhew, L.T. Krug, N. Shakhnovsky, L. Yoo and S.H. Speck, 2004. Variable methylation of the epstein-barr virus Wp EBNA gene promoter in B-lymphoblastoid cell lines. J. Virol., 78: 14062-14065.
CrossRefDirect Link - Ellmerich, S., M. Scholler, B. Duranton, F. Gosse, M. Galluser, J.P. Klein and F. Raul, 2000. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis, 21: 753-756.
PubMed - Esteller, M., M.F. Fraga, M. Guo, J. Garcia-Foncillas and I. Hedenfalk et al., 2001. DNA methylation patterns in hereditary human cancer mimics sporadic tumorigenesis. Hum. Mol. Genet., 10: 3001-3007.
Direct Link - Esteller, M. and J.G. Herman, 2002. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J. Pathol., 196: 1-7.
PubMed - Fraga, M.F., E. Uriol, L.B. Diego, M. Berdasco, M. Esteller, M.J. Canal and R. Rodriguez, 2002. High performance capillary electrophoretic method for the quantification of 5-methyl 2'-deoxycytidine in genomic DNA: Application to plant, animal and human cancer tissues. Electrophoresis, 23: 1677-1681.
PubMed - Glasspool, R.M., J.M.Teodoridis and R. Brown, 2006. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br. J. Cancer, 94: 1087-1092.
CrossRef - Gold, J.S., S. Bayar and R.R. Salem, 2004. Association of Streptococcus bovis bacteremia with colonic neoplasia and extracolonic malignancy. Arch. Surg., 139: 760-765.
PubMed - Guil, S. and M. Esteller, 2009. DNA methylomes, histone codes and miRNAs: Tying it all together. Int. J. Biochem. Cell Biol., 41: 87-95.
PubMed - Heindel, J.J., K.A. McAllister, L. Worth and F.L. Tyson, 2006. Environmental epigenomics, imprinting and disease susceptibility. Epigenetics, 1: 1-6.
Direct Link - Herman, J.G., J.R. Graff, S. Myohanen, B.D. Nelkin and S.B. Baylin, 1996. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA., 93: 9821-9826.
CrossRefDirect Link - Hermann, A., H. Gowher and A. Jeltsch, 2004. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci., 61: 2571-2587.
PubMed - Holliday, R., 1990. Mechanisms for the control of gene activity during development. Biol. Rev., 65: 431-471.
Direct Link - Hotchkiss, R.D., 1948. The quantitative separation of purines, pyrimidines and nucleotides by paper chromatography. J. Biol. Chem., 175: 315-332.
Direct Link - Huang, S., J.Y. Li, J. Wu, L. Meng and C.C. Shou, 2001. Mycoplasma infections and different human carcinomas. World J. Gastroenterol., 7: 266-269.
PubMed - Hussain, S.P., L.J. Hofseth and C.C. Harris, 2001. Tumor suppressor genes: At the crossroads of molecular carcinogenesis, molecular epidemiology and human risk assessment. Lung Cancer, 34: S7-S15.
PubMed - Ibrahim, M.A., N. Saleh, E. Archoukieh, H.W. Al-Obaide, M.M. Al-Obaidi and H.M. Said, 2010. Detection of novel genomic polymorphism in acute lymphoblastic leukemia by random amplified polymorphic DNA analysis. Int. J. Cancer Res., 6: 19-26.
CrossRef - Ibrahim, M.A., N. Saleh, K.M. Mousawy, N. Al-Hmoud, E. Archoukieh, H.W. Al-Obaide and M.M. Al-Obaidi, 2009. Molecular analysis of RAPD-PCR genomic patterns in age related acute myeloid leukemia. Trends Med. Res., 4: 35-41.
CrossRefDirect Link - Issa, J.P.J., H.M. Kantarjian and P. Kirkpatrick, 2005. Azacitidine. Nat. Rev. Drug Discovery, 4: 275-276.
Direct Link - Jablonka, E. and M.J. Lamb, 2002. The changing concept of epigenetics. Ann. N.Y. Acad. Sci., 981: 82-96.
PubMed - Johnson, T.B. and R.D. Coghill, 1925. Researches on pyrimidines. C111. The discovery of 5-methylcytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. J. Am. Chem. Soc., 47: 2838-2844.
CrossRef - Kaminskas, E., A. Farrell, S. Abraham, A. Baird and L.S. Hsieh et al., 2005. Approval summary: Azacitidine for treatment of myelodysplastic syndrome subtypes. Clin. Cancer Res., 11: 3604-3604.
Direct Link - Kaminskas, E., A.T. Farrell, Y.C. Wang, R. Sridhara and R. Pazdur, 2005. FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza™) injectable suspension. Oncologist, 10: 176-182.
PubMed - Kantarjian, H.M., S. O'Brien, J. Cortes, F.J. Giles and S. Faderl et al., 2003. Results of decitabine (5-aza-2_deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer, 98: 522-528.
PubMed - Kantarjian, H., J.P.J. Issa, C.S. Rosenfeld, J.M. Bennett and M. Albitar et al., 2006. Decitabine improves patient outcomes in myelodysplastic syndromes. Cancer, 106: 1794-1803.
Direct Link - Kastl, L., I. Brown and A.C. Schofield, 2010. Altered DNA methylation is associated with docetaxel resistance in human breast cancer cells. Int. J. Oncol., 36: 1235-1241.
PubMed - Kidwell, M.G. and D.R. Lisch, 2001. Perspective: Transposable elements, parasitic DNA and genome evolution. Evolution, 55: 1-24.
PubMed - Komashko, V.M. and P.J. Farnham, 2010. 5-azacytidine treatment reorganizes genomic histone modification patterns. Epigenetics, 5: 229-240.
Direct Link - Kornblith, A.B., J.E. Herndon, L.R. Silverman, E.P. Demakos and R. Odchimar-Reissig et al., 2002. Impact of azacytidine on the quality of life of patients with myelodysplastic syndrome treated in a randomized phase III trial: A cancer and leukemia group B study. J. Clin. Oncol., 20: 2441-2452.
PubMed - Krug, U., A. Ganser and H.P. Koeffler, 2002. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene, 21: 3475-3495.
PubMed - Kuerbitz, S.J., J. Malandro, N. Compitello, S.B. Baylin and J.R. Graff, 1999. Deletion of p16INK4A/CDKN2 and p15INK4B in human somatic cell hybrids and hybrid-derived tumors. Cell Growth Differentiation, 10: 27-33.
Direct Link - Kuehnlein, U. and W. Arber, 1972. Host specificity of DNA produced by E.coli XV. The role of nucleotide methylation in vitro B-specific modification. J. Mol. Biol., 63: 9-19.
PubMed - Lax, A.J. and W. Thomas, 2002. How bacteria could cause cancer: One step at a time. Trends Microbiol., 10: 293-299.
CrossRefDirect Link - Lind, G.E., T. Ahlquist, M. Kolberg, M. Berg and M. Eknaes et al., 2008. Hypermethylated MAL gene-a silent marker of early colon tumorigenesis. J. Transl. Med., 6: 13-13.
PubMed - Liu, L., R.C. Wylie, L.G. Andrews and T.O. Tollefsbol, 2003. Aging, cancer and nutrition: The DNA methylation connection. Mechanisms Ageing Dev., 124: 989-998.
PubMed - Luo, L., B. Li and T.P. Pretlow, 2003. DNA alterations in human aberrant crypt foci and colon Cancers by random primed polymerase chain reaction. Cancer Res., 63: 6166-6169.
PubMed - Melki, J.R., P.C. Vincent and S.J. Clark, 1999. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res., 59: 3730-3740.
Direct Link - Melki, J.R. and S.J. Clark, 2002. DNA methylation changes in leukaemia. Semin. Cancer Biol., 12: 347-357.
CrossRef - Mesa, R.A., S. Verstovsek, C. River, A. Pardanani and K. Hussein et al., 2009. 5-Azacitidine has limited therapeutic activity in myelofibrosis. Leukemia, 23: 180-182.
PubMed - Montalban, C., A. Santon, D. Boixeda and C. Bellas, 2001. Regression of gastric high grade mucosa associated lymphoid tissue (MALT) lymphoma after Helicobacter pylori eradication. Gut, 49: 584-587.
PubMed - Nyce, J., L. Liu and P.A. Jones, 1986. Variable effects of DNA-synthesis inhibitors upon DNA methylation in mammalian cells. Nucleic Acids Res., 14: 4353-4367.
PubMed - Nyce, J., 1989. Drug-induced DNA hypermethylation and drug resistance in human tumors. Cancer Res., 49: 5829-5836.
PubMed - Nguyen, C., G. Liang, T.T. Nguyen, D. Tsao-Wei and S. Groshen et al., 2001. Susceptibility of nonpromoter CpG islands to de novo methylation in normal and neoplastic cells. J. Nat. Cancer Inst., 93: 1465-1472.
PubMed - Parsonnet, J., 1995. Bacterial infection as a cause of cancer. Environ. Health Perspectives, 103: 263-268.
Direct Link - Peedicayil, J., 2008. Pharmacoepigenetics and pharcoepigenomics. Pharcogenomics, 9: 1785-1786.
PubMed - Persing, D.H. and F.G. Prendergast, 1999. Infection, immunity and cancer. Arch. Pathol. Lab. Med., 123: 1015-1022.
PubMed - Pisani, P., D.M. Parkin, N. Munoz and J. Ferlay, 1997. Cancer and infection: Estimates of the attributable fraction in 1990. Cancer Epidemiol. Biomarkers Prev., 6: 387-400.
Direct Link - Pfeifer, G.P. and T.A. Rauch, 2009. DNA methylation patterns in lung carcinomas. Semin. Cancer Biol., 19: 181-187.
CrossRef - Qiu, Y.Y., B.L. Mirkin and R.S. Dwivedi, 2005. Inhibition of DNA methyltransferase reverses cisplatin induced drug resistance in murine neuroblastoma cells. Cancer Detect. Prev., 29: 456-463.
PubMed - Radpour, R., C. Kohler, M.M. Haghighi, A.X.C. Fan, W. Holzgreve and X.Y. Zhong, 2009. Methylation profiles of 22 candidate genes in breast cancer using high-throughput MALDI-TOF mass arrayMethylation profile of breast cancer. Oncogene, 28: 2969-2978.
CrossRef - Roll, J.D., A.G. Rivenbark, W.D. Jones and W.B. Coleman, 2008. DNMT3b over expression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol. Cancer, 7: 15-15.
Direct Link - Sakai, T., J. Toguchida, N. Ohtani, D.W. Yandell, J.M. Rapaport and T.P. Dryja, 1991. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am. J. Hum. Genet., 48: 880-888.
Direct Link - Salaspuro, M.P., 2003. Acetaldehyde, microbes and cancer of the digestive tract. Crit. Rev. Clin. Lab. Sci., 40: 183-208.
PubMed - Saleh, N., M.A. Ibrahim, E. Archoukieh, A. Makkiya, M. Al-Obaidi and H. Alobydi, 2010. Identification of genomic markers by RAPD-PCR primer in leukemia patients. Biotechnology, 9: 170-175.
CrossRefDirect Link - Sanchez-Cespedes, M., M. Esteller, L. Wu, H. Nawroz-Danish and G.H. Yoo et al., 2000. Gene promoter hypermethylation in tumors serum of head and neck cancer patients. Cancer Res., 60: 892-895.
Direct Link - Santini, V., H.M. Kantarjian and J.P. Issa, 2001. Changes in DNA methylation in neoplasia: Pathophysiology and therapeutic implications. Ann. Intern. Med., 134: 573-586.
Direct Link - Saunthararajah, Y., C.A. Hillery, D. Lavelle, R. Molokie and L. Dorn et al., 2003. Effects of 5-aza-2-deoxycytidine on fetal hemoglobin levels, red cell adhesion and hematopoietic differentiation in patients with sickle cell disease. Blood, 102: 3865-3870.
PubMed - Shaw, R., 2006. The epigenetics of oral cancer. Int. J. Oral Maxillofac. Surg., 35: 101-108.
CrossRef - Schmiemann, V., A. Bocking, M. Kazimirek, A.S.C. Onofre and H.E. Gabbert et al., 2005. Methylation assay for the diagnosis of lung cancer on bronchial aspirates: A cohort study. Clin. Cancer Res., 11: 7728-7734.
Direct Link - Schmid, C.W., 1991. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res., 19: 5613-5617.
PubMed - Schmid, C.W., 1998. Does SINE evolution preclude Alu function. Nucleic Acids Res., 26: 4541-4550.
Direct Link - Shilatifard, A., 2006. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem., 75: 243-269.
Direct Link - Shin, C.M., N. Kim, Y. Jung, J.H. Park and G.H. Kang et al., 2010. Role of Helicobacter pylori infection in aberrant DNA methylation along multistep gastric carcinogenesis. Cancer Sci., 101: 1337-1346.
Direct Link - Shukla, V.K., H. Singh, M. Pandey, S.K. Upadhyay and G. Nath, 2000. Carcinoma of the gallbladder-is it a sequel of typhoid?. Dig. Dis. Sci., 45: 900-903.
PubMed - Srinivasan, P.R. and E. Borek, 1964. Enzymatic alteration of nucleic acid structure. Science, 145: 548-553.
PubMed - Strathdee, G. and R. Brown, 2002. Aberrant DNA methylation in cancer: Potential clinical interventions. Expert. Rev. Mol. Med., 4: 1-17.
Direct Link - Stirzaker, C., D.S. Millar, C.L. Paul, P.M. Warnecke and J. Harrison et al., 1997. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res., 57: 2229-2237.
PubMed - Suzuki, M. and I. Yoshino, 2008. Identification of micro RNAs caused by DNA methylation that induce metastasis. Future Oncol., 4: 775-777.
PubMed - Szalmas, A. and J. Konya, 2009. Epigenetic alterations in cervical carcinogenesis. Semin. Cancer Biol., 19: 144-152.
CrossRef - Szyf, M., P. Pakneshanb and S.A. Rabbani, 2004. DNA methylation and breast cancer. Biochem. Pharmacol., 68: 1187-1197.
Direct Link - Tucker, K.L., C. Beard, J. Dausman, L. Jackson-Grusby and P.W. Laird et al., 1996. Germ-line passage is required for establishment of methylatlon and expression patterns of imprinted but not of non imprinted genes. Genes Dev., 10: 1008-1020.
Direct Link - Vaishnavi, C., R. Kochhar, G. Singh, S. Kumar, S. Singh and K. Singh, 2005. Epidemiology of typhoid carriers among blood donors and patients with biliary, gastrointestinal and other related diseases. Microbiol. Immunol., 49: 107-112.
PubMed - Vakevainen, S., S. Mentula, H. Nuutinen, K.S. Salmela, H. Jousimies-Somer, M. Farkkila and M. Salaspuro, 2002. Ethanol-derived microbial production of carcinogenic acetaldehyde in achlorhydric atrophic gastritis. Scandinavian J. Gastroenterol., 37: 648-655.
Direct Link - Veeck, J., E. Noetzel, N. Bektas, E. Jost, A. Hartmann, R. Knuchel and E. Dahl, 2008. Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumor-specific epigenetic marker in human breast cancer. Mol. Cancer, 7: 83-83.
PubMed - Velculescu, V.E. and W.S. El-Deiry, 1996. Biological and clinical importance of the p53 tumor suppressor gene. Clin. Chem., 42: 858-868.
PubMed - De Vos, D. and W. Van Overveld, 2005. Decitabine: A historical review of the development of an epigenetic drug. Ann. Hematol., 84: 3-8.
Direct Link - Wang, C., B.L. Mirkin and R.S. Dwivedi, 2001. DNA (cytosine) methyltransferase overexpression is associated with acquired drug resistance of murine neuroblastoma cells. Int. J. Oncol., 18: 323-329.
PubMed - Waterland, R.A. and K.B. Michels, 2007. Epigenetic epidemiology of the developmental origins hypothesis. Annu. Rev. Nutr., 27: 363-388.
PubMed - Yang, A.S., M.R.H. Esteacio, K. Doshi, Y. Kondo, E.H. Tajara and J.J. Issa, 2004. A simple method for estimating global DNA methylation using bisulfte PCR of repetitive DNA elements. Nucleic Acids Res., 32: e38-e38.
Direct Link - Zarkin, B.A., K.D. Lillemoe, J.L. Cameron, P.N. Effron, T.H. Magnuson and H.A. Pitt, 1990. The triad of Streptococcus bovis bacteremia, colonic pathology and liver disease. Ann. Surg., 211: 786-791.
PubMed - Zhu, J. and X. Yao, 2009. Use of DNA methylation for cancer detection: Promises and challenges. Int. J. Biochem. Cell Biol., 41: 147-154.
CrossRef - Perez-Plasencia, C. and A. Duenas-Gonzalez, 2006. Can the state of cancer chemotherapy resistance be reverted by epigenetic therapy? Mol. Cancer, Vol. 5.
CrossRefDirect Link - Segura-Pacheco, B., E. Perez-Cardenas, L. Taja-Chayeb, A. Chavez-Blanco, A. Revilla-Vazquez, L. Benitez-Bribiesca and A. Duenas-Gonzalez, 2006. Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J. Transl. Med., Vol. 4.
CrossRefDirect Link









ANAMIKA BARMAN Reply
Really I find interest in your abstract.But sir I want to know what are those bacteria which change the pattern of DNA.I am M.Sc in zoology with specialization in Animal Physiology and Biochemistry with 72 percent.
Mohammed A. Ibrahim
Dear Barman,
Thanks for your interest, open the fulltext PDF and have a look at table 3 you will find the bacteria and the references, they include: H. pylori, Salmonella typhi, Mycoplasma and Streptococcus bovis.
Regards
M. Ibrahim