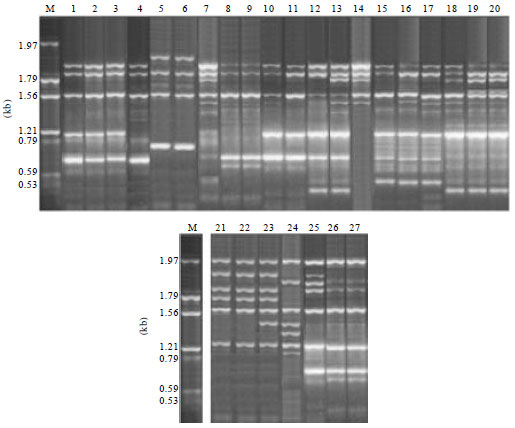
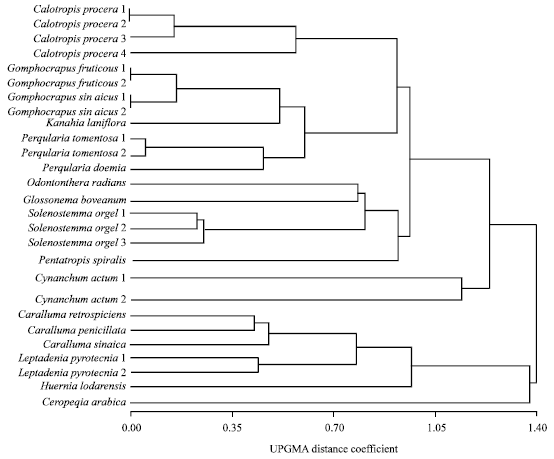
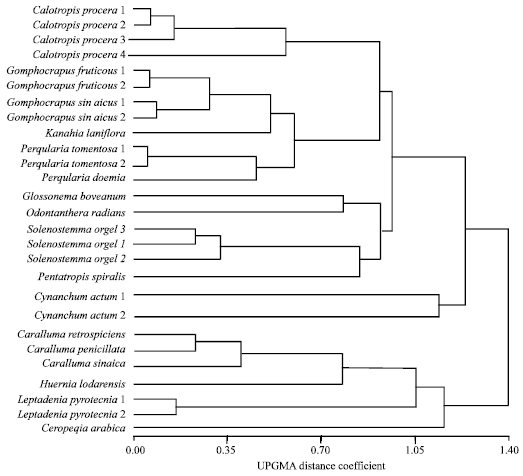
Research Article
Taxonomic Relationships of Some Taxa of Subfamily Asclepiadoideae (Apocynaceae) as Reflected by Morphological Variations and Polymorphism in Seed Protein and RAPD Electrophoretic Profile
Department of Botany, Faculty of Science, Ain Shams University, Abbaseya 11566, Cairo, Egypt