Research Article
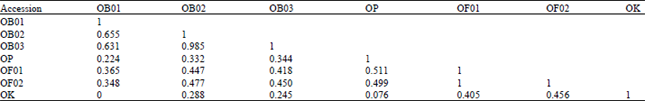
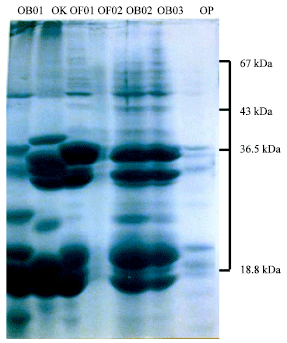
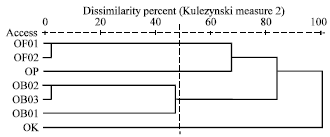
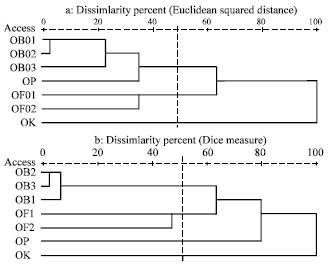
Genetic Diversity among Ocimum Populations in Egypt as Reflected by Morphological, Seed Proteins and Isozyme Polymorphism
Department of Botany, Faculty of Science, Tanta University, 31527 Tanta, Egypt
Abdelfattah Badr
Department of Botany, Faculty of Science, Tanta University, 31527 Tanta, Egypt
Mohammed A. El-Galaly
Department of Botany, Faculty of Science, Benha University, Benha, Egypt
Ahmed A. Mobarak
Department of Botany, Faculty of Science, Benha University, Benha, Egypt
Mervet G. Hassan
Department of Botany, Faculty of Science, Benha University, Benha, Egypt