Research Article
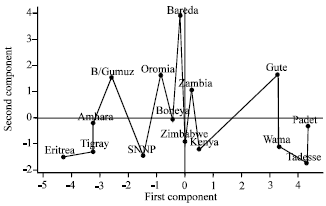
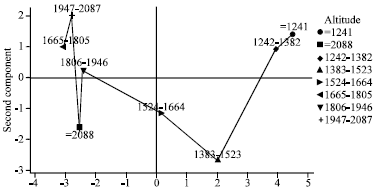
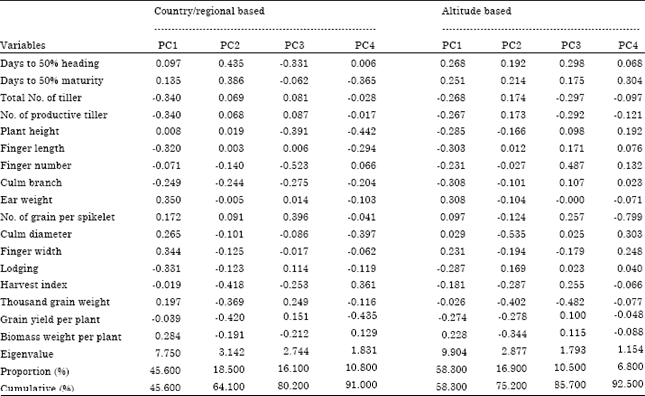
Multivariate Analysis for Quantitative Traits in Finger Millet (Eleusine coracana subsp. coracana) Population Collected from Eastern and Southeastern Africa: Detection for Patterns of Genetic Diversity
Bako Agricultural Research Center, P.O. Box 03, Bako, Ethiopia
Kassahun Tesfaye
Faculty of Life Science, Addis Abba University, Addis Abba, Ethiopia
Masresha Fetene
Faculty of Life Science, Addis Abba University, Addis Abba, Ethiopia
Santie de Villiers
Faculty of Life Science, Addis Abba University, Addis Abba, Ethiopia