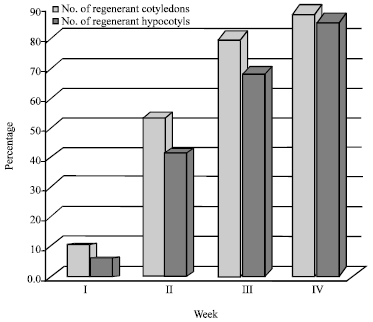
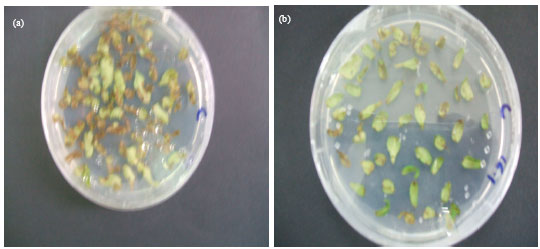
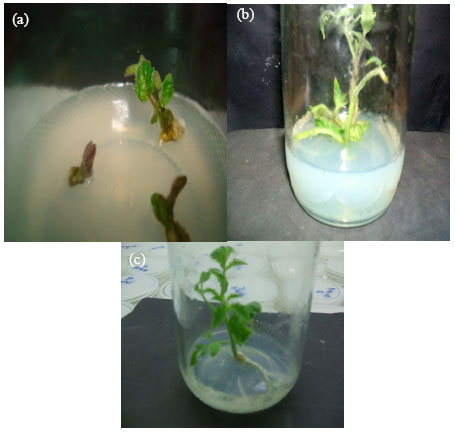
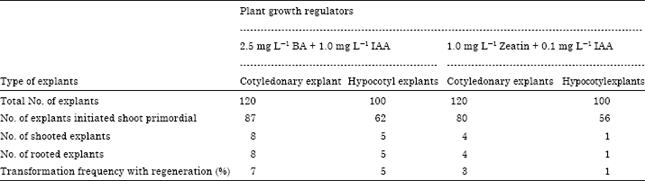
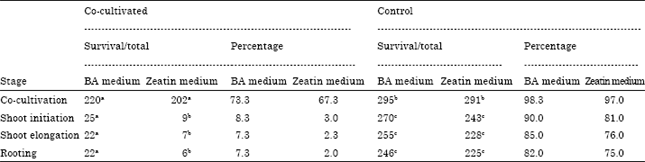
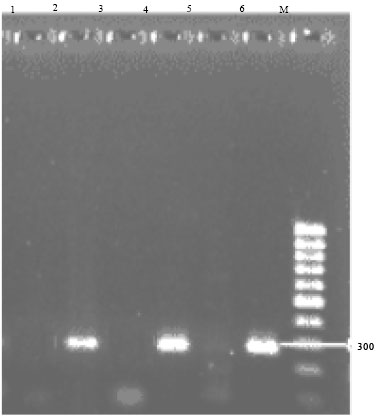
Research Article
Agrobacterium-mediated Transformation of Tomato Plants Expressing Defensin Gene
Depertment of Botany, Faculty of Science, University of Khartoum, P.O. Box 321, P.C. 11115, Sudan
A.A. El-Hussein
Depertment of Botany, Faculty of Science, University of Khartoum, P.O. Box 321, P.C. 11115, Sudan
M.M. Saker
Academy of Scientific Research and Technology, Egypt