Research Article
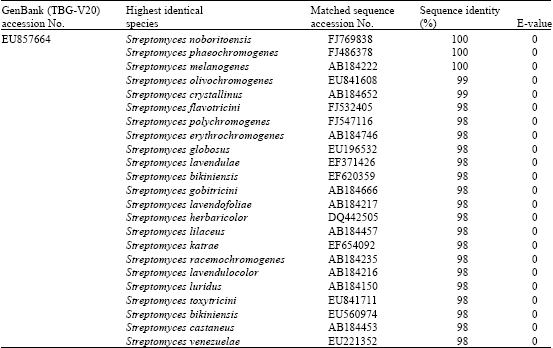
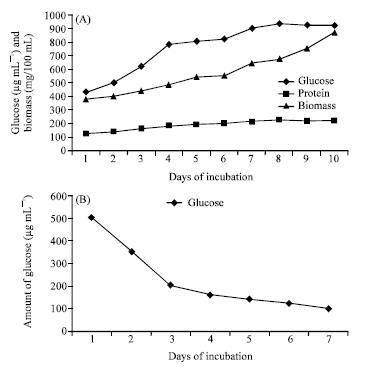
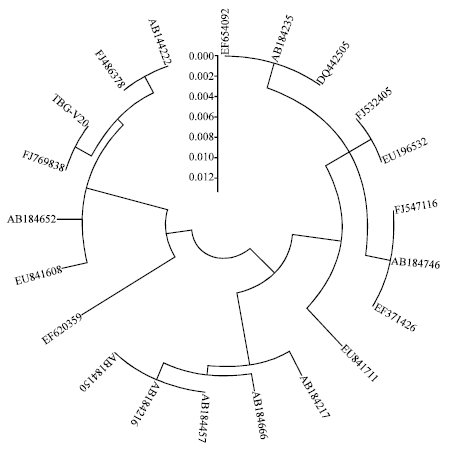
Novel Approaches for Identification of Streptomyces noboritoensis TBG-V20 with Cellulase Production
Environmental Nanotechnology Division, Sri Paramakalyani Centre for Environmental Sciences, Manonmaniam Sundaranar University, Alwarkurichi-627 412, Tamil Nadu, India
E.G. Wesely
Centre for Biotechnology, Muthayammal College of Arts and Science, Rasipuram-637408, Tamil Nadu, India
J. George
Centre for Biotechnology, Muthayammal College of Arts and Science, Rasipuram-637408, Tamil Nadu, India
G. Annadurai
Environmental Nanotechnology Division, Sri Paramakalyani Centre for Environmental Sciences, Manonmaniam Sundaranar University, Alwarkurichi-627 412, Tamil Nadu, India