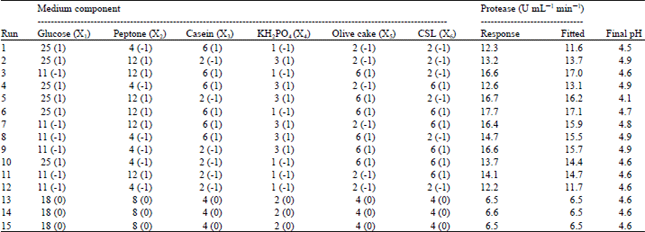
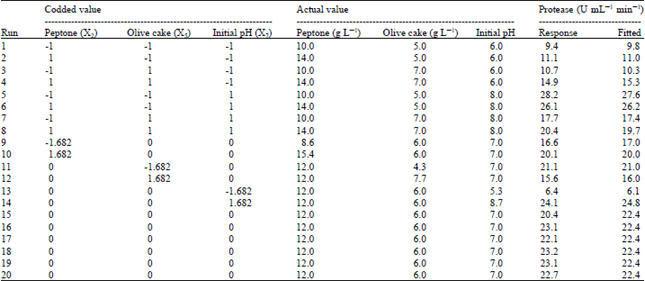
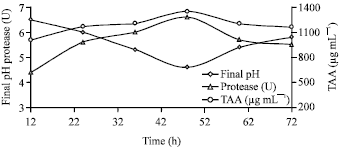
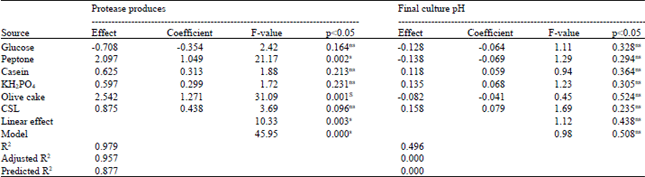
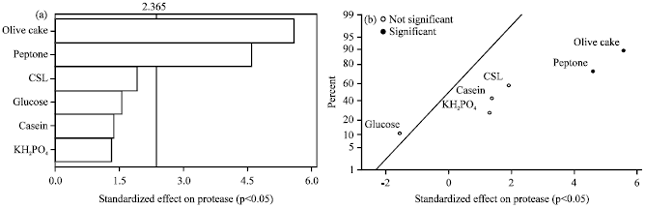
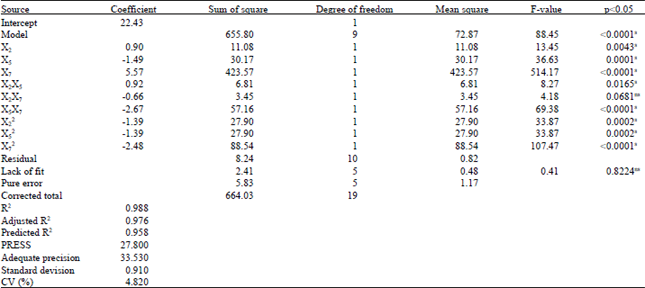
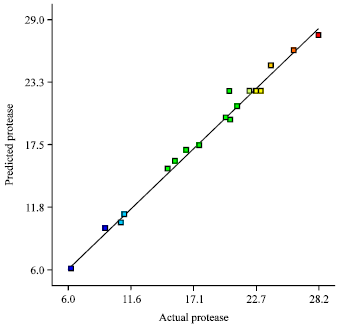
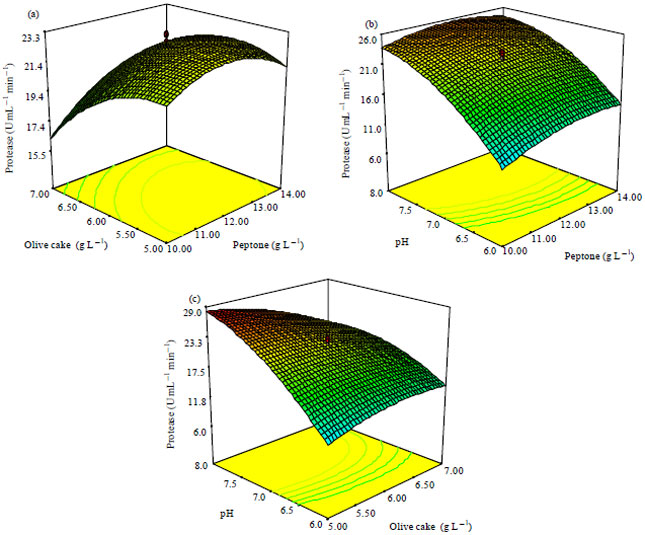
Research Article
Amino Acids Associated with Optimized Alkaline Protease Production by Bacillus subtilis ATCC 11774 Using Statistical Approach
Microbial Activity Unit, Department of Microbiology, Soils, Water and Environment Research Institute, Agricultural Research Center, P.N. 12619, Giza, Egypt
Wesam I.A. Saber
Microbial Activity Unit, Department of Microbiology, Soils, Water and Environment Research Institute, Agricultural Research Center, P.N. 12619, Giza, Egypt
Husain A. El-Fadaly
Department of Microbiology, Faculty of Agriculture, Damietta University, Egypt