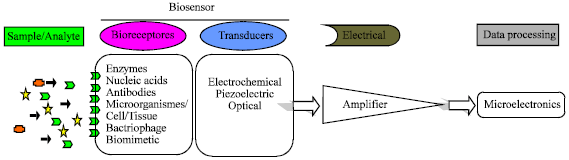
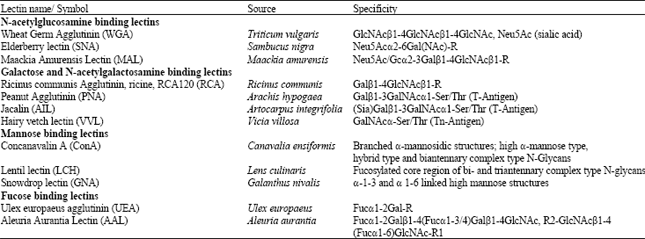
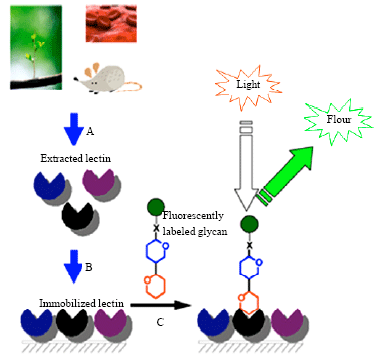
Review Article
Lectin-based Biosensors: As Powerful Tools in Bioanalytical Applications
Biotechnology Research Institute, School of Agriculture, Shiraz University, Shiraz, Iran
S.S. Kazemi
Department of Plant Breeding, Faculty of Agricultural Sciences and Engineering, College of Agriculture and Natural Resources, University of Tehran, Karaj, Iran