Research Article
In Silico Analysis of Evolution in Swine Flu Viral Genomes Through Re-assortment by Promulgation and Mutation
NBU Bioinformatics Faculty, University of North Bengal, Siliguri 734013, India
G. Sen
NBU Bioinformatics Faculty, University of North Bengal, Siliguri 734013, India
S. Thakur
NBU Bioinformatics Faculty, University of North Bengal, Siliguri 734013, India
A.K. Bothra
Department of Chemistry, Raiganj College, Raiganj 733134, India
A. Sen
NBU Bioinformatics Faculty, University of North Bengal, Siliguri 734013, India










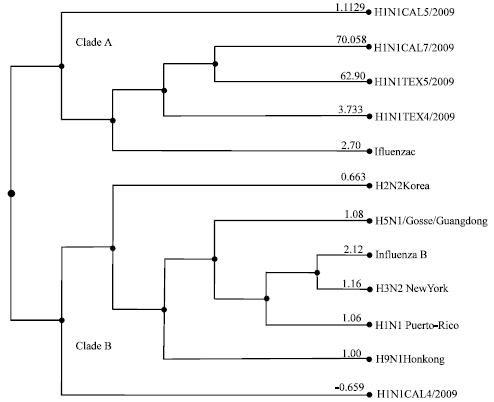
shigha t Reply
Is this H1N1 virus is influensed by anyother viruses
Saubashya Sur
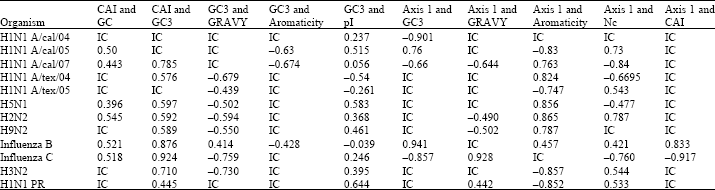
It has been found that HIN1 viral strains are not influenced by any other viruses. We studied the neuraminidase and hemagglitinin genes from H1N1 viruses and found that other viruses do not influence them.