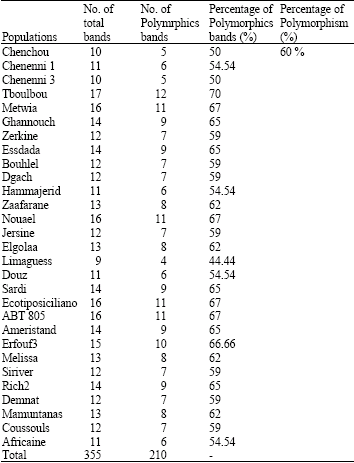
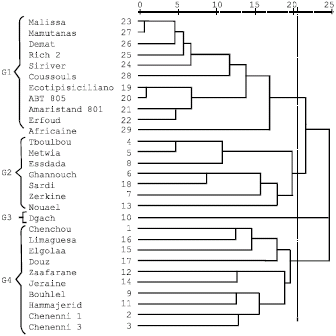
Research Article
Genetic Diversity of Some Mediterranean Populations of the Cultivated Alfalfa (Medicago sativa L.) Using ISSR Markers
Institute of the Arids Regions, Rue Eljorf, km 22.5 Medenine 4119, Tunisia
F. Guesmi
Institute of the Arids Regions, Rue Eljorf, km 22.5 Medenine 4119, Tunisia
K. Fares
Institute of the Arids Regions, Rue Eljorf, km 22.5 Medenine 4119, Tunisia
A. Ferchichi
Institute of the Arids Regions, Rue Eljorf, km 22.5 Medenine 4119, Tunisia