Research Article
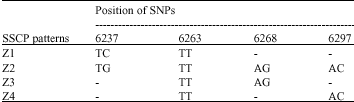
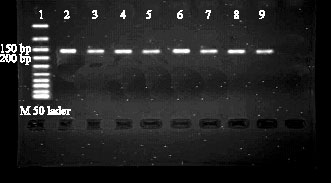
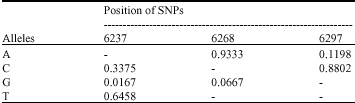
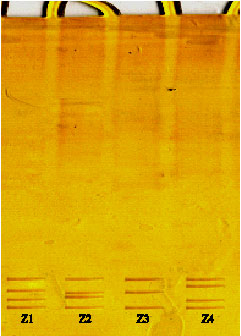
Characterization of SNPs of Bovine Prolcatin Gene of Holstein Cattle
Department of Animal Science, Faculty of Agriculture, University of Zanjan, Zanjan, Iran
Morad Pasha Eskandari Nasab
Department of Animal Science, Faculty of Agriculture, University of Zanjan, Zanjan, Iran
Mohammad Reza Nassiry
Center of Excellence in Animal Science, Faculty of Agriculture, Ferdowsi University of Mashhad, Mashhad, Iran
Ali Reza Heravi Mossavi
Center of Excellence in Animal Science, Faculty of Agriculture, Ferdowsi University of Mashhad, Mashhad, Iran
Seyed Abolfazl Hosseini
Department of Biology, Faculty of Science, STT University, Sabzevar, Iran
Saber Qanbari
Department of Animal Science, Faculty of Agriculture, University of Zanjan, Zanjan, Iran