Research Article
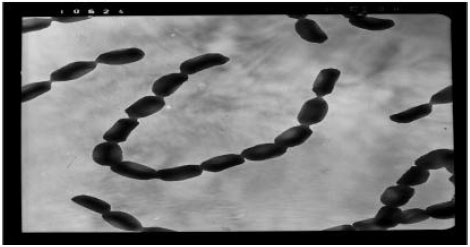
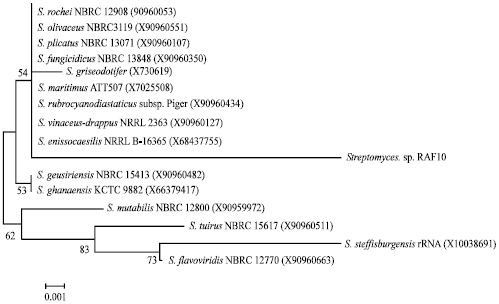
Screening, Isolation and Characterization of a Novel Antimicrobial Producing Actinomycete, Strain RAF10
Department of Microbiology, Faculty of Sciences, Houari Boumediene University of Science and Technology, Bab-Azzouar, Algiers-Algeria
Ali Elshafei
Department of Microbial Chemistry, National Research Centre (NRC), Dokki, Cairo, Egypt
Mahmoud Saker
Plant Molecular Genetics Group, Nobel Project, National Research Centre (NRC), Dokki, Cairo, Egypt
Bengraa Cheikh
Industrial Chemistry Department, Faculty of Sciences, University of Science and Technology, Oran-Algeria
Hacene Hocine
Department of Microbiology, Faculty of Sciences, Houari Boumediene University of Science and Technology, Bab-Azzouar, Algiers-Algeria