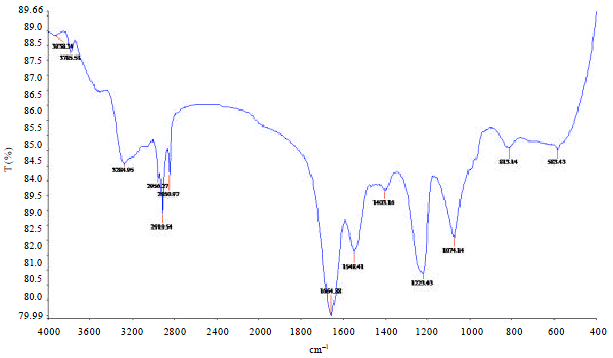
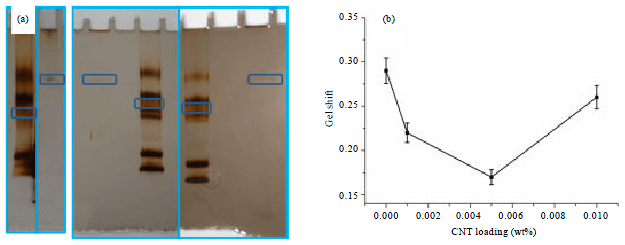
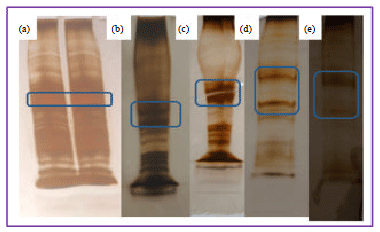
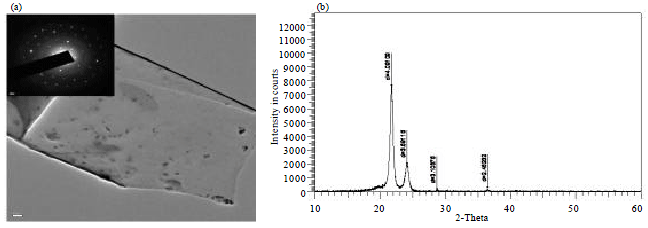
Research Article
Lotus-leaf Inspired Hydrophobic Nanocomposite Matrices for Electrophoretic Separation of Bacterial Outer Membrane Proteins
Centre for Nanotechnology and Advanced Biomaterials (CeNTAB), School of Chemical and Biotechnology, SASTRA University, 613401, Thanjavur, India
Vidya N. Chamundeswari
Centre for Nanotechnology and Advanced Biomaterials (CeNTAB), School of Chemical and Biotechnology, SASTRA University, 613401, Thanjavur, India
Meera Parthasarathy
Centre for Nanotechnology and Advanced Biomaterials (CeNTAB), School of Chemical and Biotechnology, SASTRA University, 613401, Thanjavur, India