Research Article
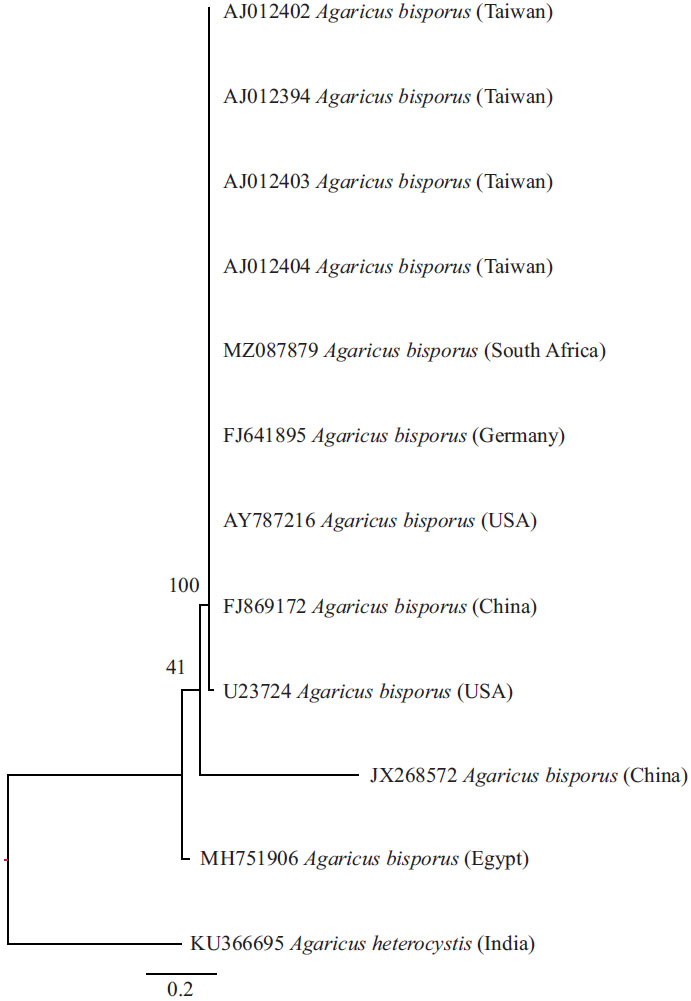
Natural Strain Observation of Button Mushroom (Agaricus bisporus) from South Africa Using 18S rDNA
Aquaculture Research Unit, Faculty of Science and Agriculture, School of Agricultural and Environmental Sciences, University of Limpopo, Turfloop Campus, Private Bag X1106, Sovenga 0727, South Africa
LiveDNA: 98.36021
ORCID: 0000-0002-3847-5648