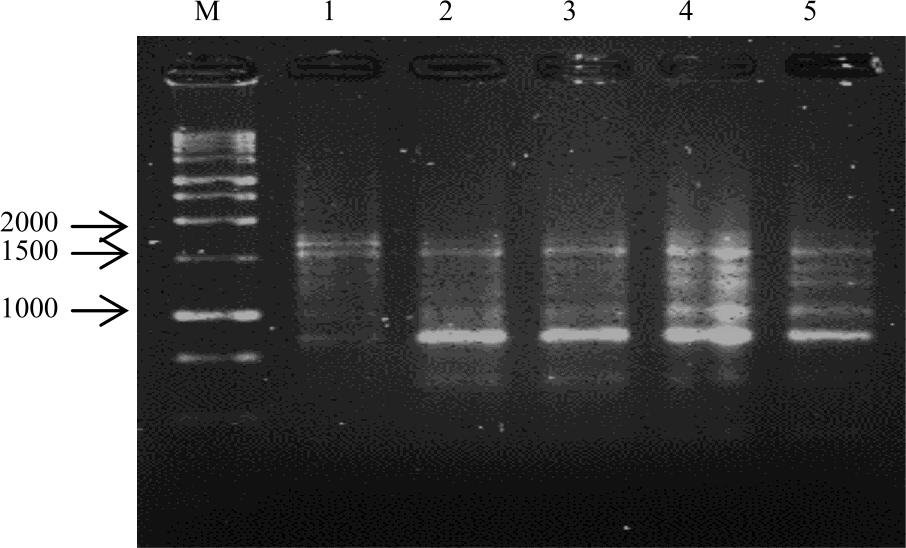
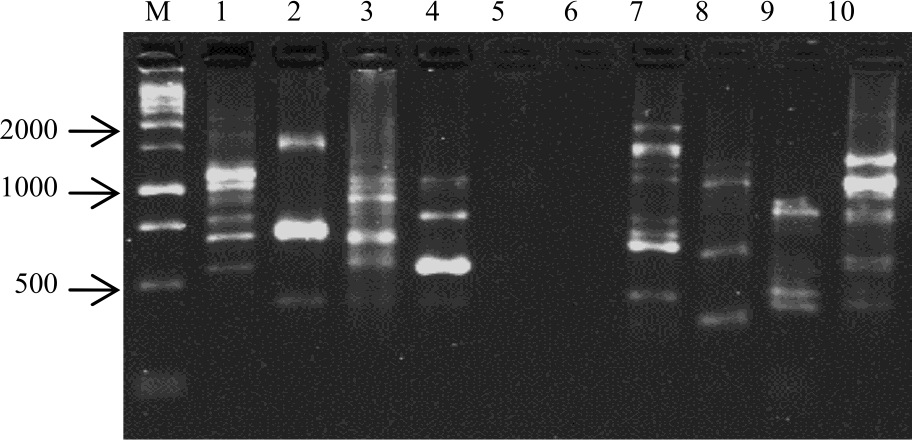
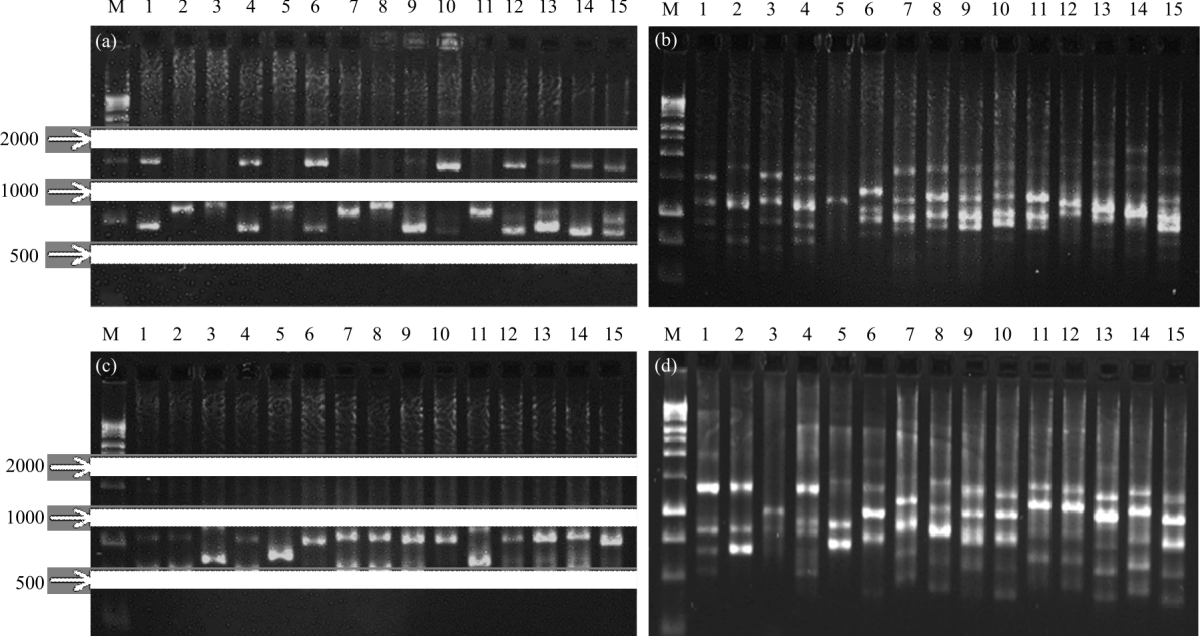
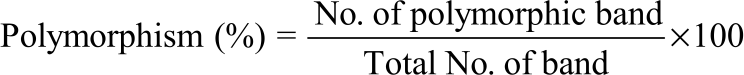Research Article
Genetic Diversity of Threatened Aquilaria sp. in Malaysia Using Inter Simple Sequence Repeat (ISSR) Markers
Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Kampus Besut, 22200 Besut, Terengganu, Malaysia
Hasan Nudin Nur Fatihah
Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Kampus Besut, 22200 Besut, Terengganu, Malaysia
LiveDNA: 60.31303
Syaidatul Najiah Zakaria
Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Kampus Besut, 22200 Besut, Terengganu, Malaysia
Nadiawati Alias
Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Kampus Besut, 22200 Besut, Terengganu, Malaysia
Nor Hasima Mahmod
Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Kampus Besut, 22200 Besut, Terengganu, Malaysia