Research Article
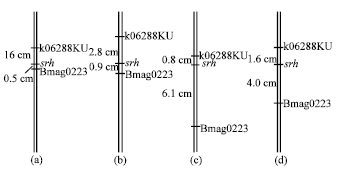
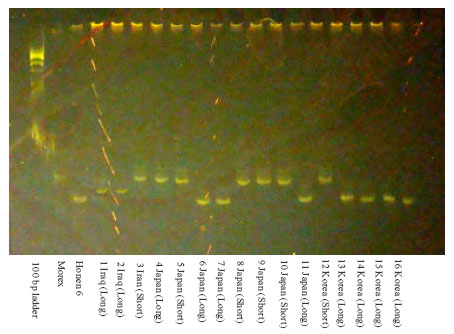
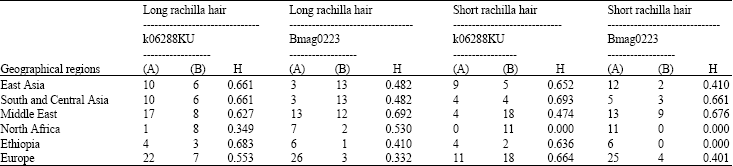
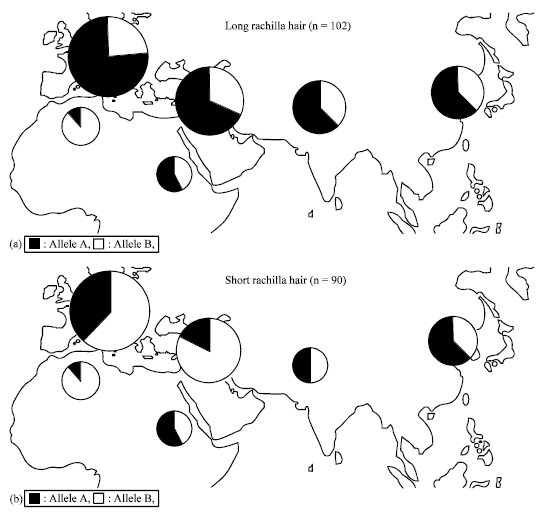
Fine Mapping of Short Rachilla Hair Gene (srh) in Barley and an Association Study using Flanking Molecular Markers and World Germplasms
Faculty of Agriculture, Kagawa University, Ikenobe, Miki Cho, Kita Gun, Kagawa 761-0795, Japan
S. Ikeda
Faculty of Agriculture, Kagawa University, Ikenobe, Miki Cho, Kita Gun, Kagawa 761-0795, Japan
I. Kataoka
Faculty of Agriculture, Kagawa University, Ikenobe, Miki Cho, Kita Gun, Kagawa 761-0795, Japan
S. Taketa
Research Institute for Bioresources, Okayama University, Kurashiki 710-0046, Japan