Research Article
Identification and Differentiation of Three Nigerian Orseolia sp. by RAPD Markers
Not Available
A. Onasanya
Not Available
O. Okhidievbie
Not Available
Y. Sere
Not Available
M.N. Ndjiondjop
Not Available
I. Ingelbrecht
Not Available
The African Rice Gall Midge (AfRGM), Orseolia oryzivora Harris and Gagné (Diptera: Cecidomyiidae), is an insect pest of rainfed and irrigated lowland rice in Nigeria and has been especially troublesome during the past 15 years as rice production has intensified. It is widely distributed in Burkina Faso[1,2], Mali (Hamadoun, personal communication), Nigeria[3-6], Sierra Leone[7] and Cameroon[8] and has also been recorded in a further 16 sub-Saharan African countries[9]. Orseolia bonzii, which develops on the common weed, Paspalum scrobiculatum, is a distinct species which cannot attack rice but is an alternative host for at least one of the two main parasitoids of AfRGM[5]. The O. nwanzei develops on Eragrostis atrovirens (Desf.) Trin. ex Steud, which is a widespread weed of rice crops, especially in the savanna and derived savanna zones of Nigeria.
Although the three Orseolia species exist in Nigeria, it is very difficult or impossible to identify and differentiate them based on morphological characters. The present study aims to circumvent these problems by developing a DNA-based technique for accurate and rapid identification of species (named and unnamed). Frequently-used DNA analytical techniques are based on Restriction Fragment Length Polymorphisms (RFLPs)[10] and specific amplification of polymorphic DNA fragments by PCR[11,12]. But the intensive labor and time inputs[13] and the need for effective probes often make RFLP analysis cumbersome. Use of the Polymerase Chain Reaction (PCR) facilitates detection of Random Amplified Polymorphic DNAs (RAPDs) using arbitrarily designed short primers to amplify DNA[14]. This PCR-RAPD technique has been previously employed to detect DNA polymorphisms in insects[15-17] and has the advantage that no previous sequence information or probe identity is needed for the fingerprinting. Accurate identification of the various species of Orseolia is therefore of great practical importance, especially in plant breeding programs but also in the development of IPM.
Insect species: Adults and pupae of Orseolia nwanzei sp. n., Orseolia oryzivora and Orseolia. bonzii used in this study were obtained in September 2004 from the Entomology Unit, WARDA, Ibadan, Nigeria. After artificial rearing in 2004, insects were preserved at –20°C in absolute ethanol before DNA extraction.
DNA extraction: DNA extraction of 15 adult insects and 15 pupae (Table 1) were carried out[11,18]. Whole insects were ground in liquid nitrogen, suspended in 200 μL of 2xCTAB buffer (50 mM Tris, pH 8.0; 0.7 mM NaCl; 10 mM EDTA; 2% hexadecyltrimethylammonium bromide; 0.1% 2-mercaptoethanol), followed by the addition of 50 μL of 20% sodium dodecyl sulfate and incubated at 65°C for 30 min. DNA was purified by two extractions with phenolchloroformisoamyl alcohol (24:25:1) and precipitated with -20°C absolute ethanol.
| Table 1: | Identity of insect species used in this study |
 | |
| Table 2: | Oligonucleotide primers that showed genetic discrimination among the Orseolia sp. using RAPD-PCR analysis |
 | |
After washing with 70% ethanol, the DNA was dried and re-suspended in 150 μL of sterile distilled water. DNA concentration was measured using a DU-65UV spectrophotometer (Beckman Instruments Inc., Fullerto CA, USA) at 260 nm. DNA degradation was checked by electrophoresis on a 1% agarose gel in 1xTAE (45 mM Tris-acetate, 1 mM EDTA, pH 8.0).
RAPD-PCR analysis: RAPD-PCR analysis was conducted on DNA of the three Orseolia sp.[19]. DNA primers tested were purchased from Operon Technologies (Alameda, California, USA) and each was 10 nucleotides long. Two concentrations of each DNA (25 and 95 ng per reaction) were used to test reproducibility and eliminate sporadic amplification products from the analysis. Ninety primers (OPP, OPQ, OPR, OPS, OPT, OPV, OPX and OPY series) were screened with DNA of Orseolia nwanzei, Orseolia bonzii and Orseolia oryzivora for their ability to amplify the insect DNA. Six of these primers (Table 2) were found useful since they gave polymorphism. These were used in amplifying the DNA from all the adults and pupae. Amplifications were performed in 25 μL reaction mixture consisting of genomic DNA, 1X reaction buffer (Promega), 100 μM each of dATP, dCTP, dGTP and dTTP, 0.2 μM Operon random primer, 2.5 μM MgCl2 and 1U of Taq polymerase (Boehringer, Germany). A single primer was used in each reaction. The reaction mixture was overlaid with 50 μL of mineral oil to prevent evaporation. Amplification was performed in a thermowell microtiter plate (Costa Corporation) using Perkin Elmer programmable Thermal Controller model 9600. The cycling program was (I) 1 cycle of 94°C for 3 min; (ii) 45 cycles of 94°C for 1 min for denaturation, 40°C for 1 min for annealing of primer and 72°C for 2 min for extension and (iii) a final extension at 72°C for 7 min. The amplification products were resolved by electrophoresis in a 1.4% agarose gel using TAE buffer (45 mM Tris-acetate, 1 mM EDTA, pH 8.0) at 100 V for 2 h. A 1 kb ladder (Life Technologies, Gaithersburg, MD, USA) was included as molecular size marker. Gels were visualized by staining with ethidium bromide solution (0.5 μg mL-1) and banding patterns were photographed over UV light using a red filter.
Cluster analysis: Positions of unequivocally scorable RAPD bands were transformed into a binary character matrix (1 for the presence and 0 for the absence of a band at a particular position). Pairwise distance matrices were compiled by the NTSYS-pc 2.0 software[20] using the Jaccard coefficient of similarity[21]. A dendrogram was created by the unweighted pair-group method arithmetic (UPGMA) average cluster analysis[22,23].
After screening of 90 primers with DNA of Orseolia nwanzei, Orseolia bonzii and Orseolia oryzivora, only 6 primers (OPR-04, OPS-07, OPT-08, OPX-20, OPV-06, OPV-16) gave reproducible polymorphism and useful genetic information that differentiates the three insect species. Amplification with the 6 primers generated 54 bands, 37 (69%) of them being polymorphic (Table 2) with sizes ranging between 1000 and 3000 base pairs (Fig. 1). For each insect species, DNA fingerprints of five adult insects and five pupae were found to be genetically the same and were not influenced by stage of insect development (Fig. 1). Consequently, a bulk DNA of adults (5 insects) and a bulk DNA of pupae (5 pupae) were genetically the same for each insect species (Fig. 1). Using 37 RAPD markers (Table 2) in cluster analysis at species level revealed that Orseolia nwanzei, Orseolia oryzivora and Orseolia. bonzii are genetically distinct (Fig. 2).
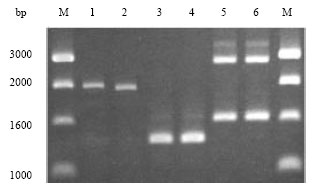 | |
| Fig. 1: | DNA fingerprinting patterns of Orseolia oryzivora, O. bonzii and O. nwanzei sp. n. using OPS-7 RAPD primer. M: 1kb molecular size marker. 1: O. oryzivora pupae, 2: O. oryzivora adults, 3: O. bonzii pupae, 4: O. bonzii adults, 5: O. nwanzei pupae, 6: O. nwanzei adults |
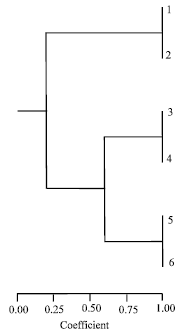 | |
| Fig. 2: | Cluster analysis of Orseolia species as revealed by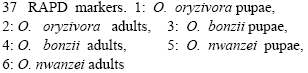 |
With a genetic similarity coefficient of about 60%, Orseolia oryzivora and Orseolia bonzii share a common genetic origin, leaving Orseolia nwanzei distinct from those two species (Fig. 2).
The existence of genetic variation between the three Orseolia sp., as revealed by RAPDs markers, demonstrates its fingerprinting and diagnostic potential that could be used to complement morphological characters[24,25]. The use of genetic fingerprinting in management of insects strongly supports the effort to efficiently and effectively identify genetic variation in the species. Insect classification and genetic relationships are important issues for entomologists working on host plant resistance and biological control. The application of RAPDs seems very useful in this regard[25]. Using RAPD to determine genetic relationships should therefore allow entomologists to identify and differentiate insect species before release in the field. This will also assist the scientists to study existing species composition in a locality before release of a new species, thereby detecting the level of outcross between other species in the field.
The DNA fingerprint defined by each Orseolia sp. should be useful for surveys, identification of new species and to differentiate aggressive from non-aggressive species. This information will strongly assist breeding programs aiming at the effective development of cultivars with durable resistance to AfRGM.
The authors are grateful to the technical staff of the Central Biotechnology Laboratory, International Institute of Tropical Agriculture for RAPD-PCR analysis.