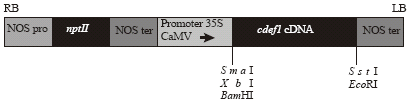
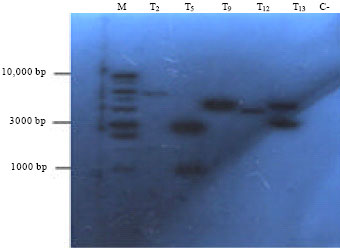
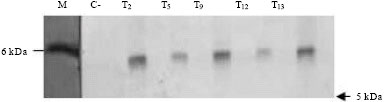
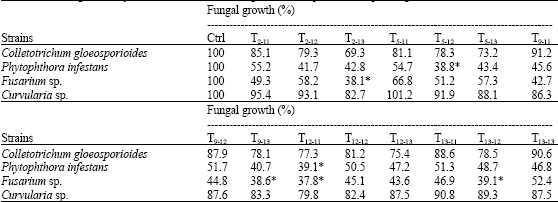
Research Article
Expression of the Capsicuum annum (Chili) Defensin Gene in Transgenic Tomatoes Confers Enhanced Resistance to Fungal Pathogens
School of Biosciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor Darul Ehsan, Malaysia
E. Marouf
School of Biosciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor Darul Ehsan, Malaysia
I. Ismail
School of Biosciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor Darul Ehsan, Malaysia
C.K. Fei
School of Biosciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Selangor Darul Ehsan, Malaysia