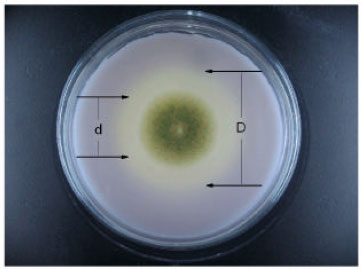
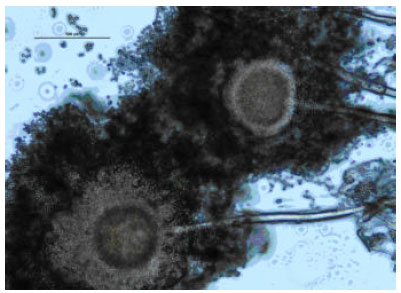
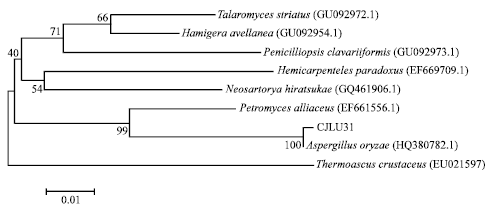
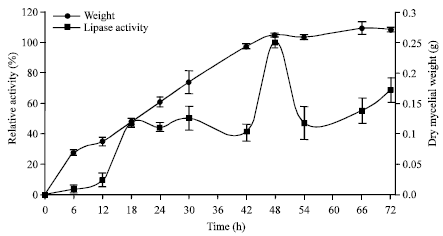
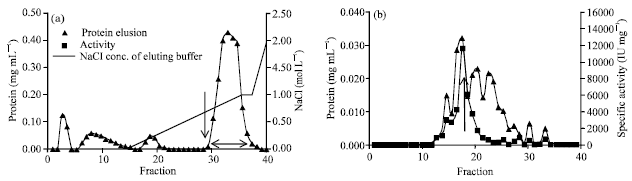
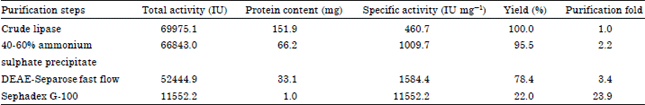
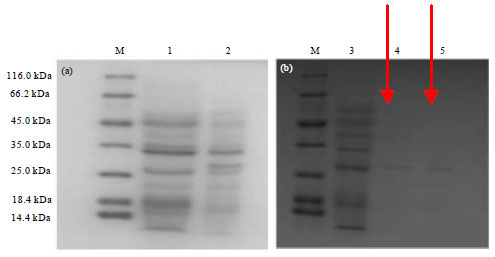
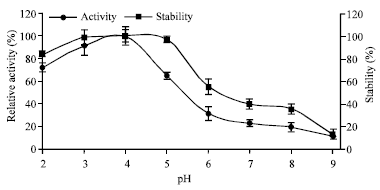
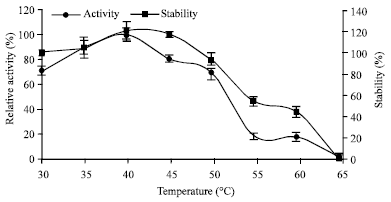
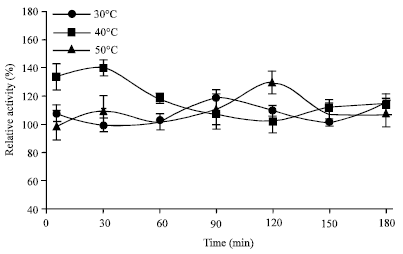
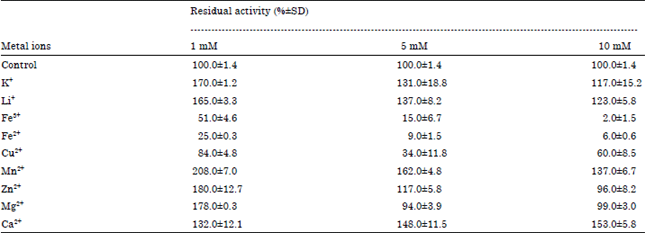
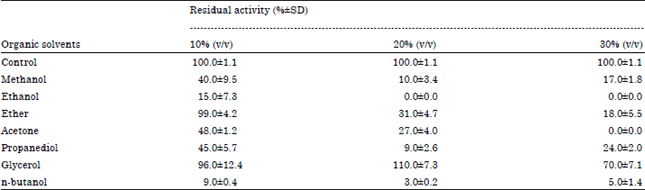
Research Article
Purification and Characterization of Lipase Produced by Aspergillus oryzae CJLU-31 Isolated from Waste Cooking Oily Soil
College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, China
Wen-Wei Chen
College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, China
Zheng-Bao Jia
College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, China
Guang-Rong Huang
College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, China
Yao Hong
College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, China
Jun-Jie Tao
College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, China
Xiao-Bo Luo
College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, 310018, China