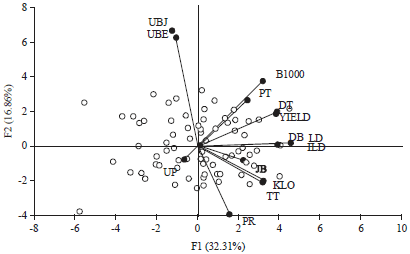
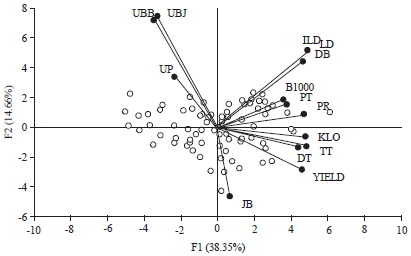
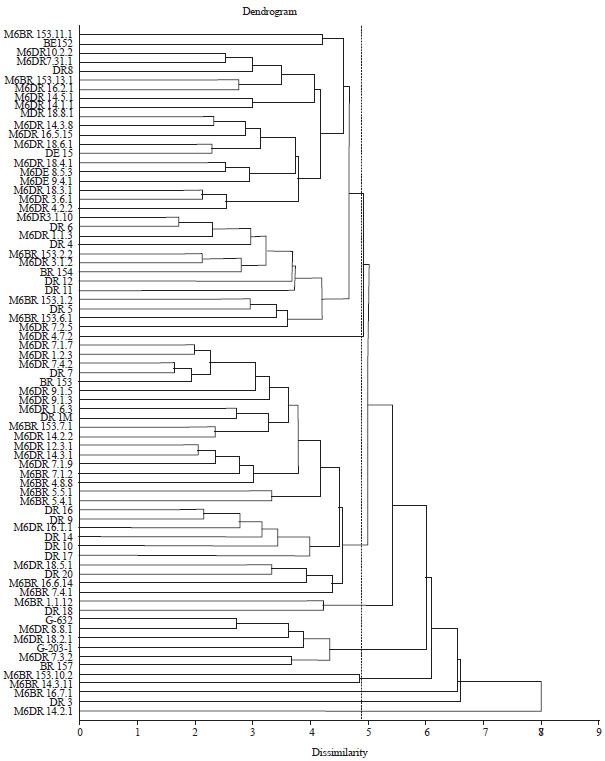
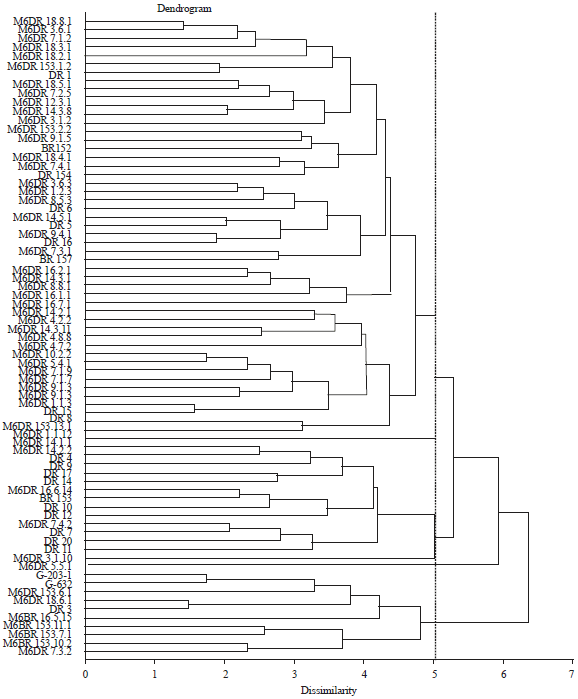
Research Article
Multivariate Analysis of Genetic Diversity among some Maize Genotypes under Maize-Albizia Cropping System in Indonesia
Faculty of Agriculture, Universitas Singa Perbangsa, Karawang, Indonesia
I. Cartika
Faculty of Agriculture, Universitas Majalengka, Indonesia
D. Ruswandi
Padjadjaran University, Bandung, 40600, Indonesia