Research Article
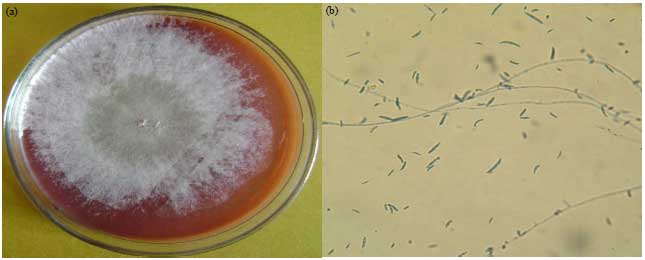
Occurrence and Prevalence of Mycotoxigenic Fusarium solani in Onion Samples Collected from Different Regions of Libya
Center for Global Programs (CGP), Management and Science University (MSU), Shah Alam, Selangor, Malaysia
Mohammed Abdelfatah Alhoot
International Medical School (IMS), Management and Science University (MSU), University Drive, Off Persiaran Olahraga, Seksyen 13, Shah Alam, 40100, Selangor Darul Ehsan, Malaysia
LiveDNA: 970.24768
Kartikeya Tiwari
International Medical School (IMS), Management and Science University (MSU), University Drive, Off Persiaran Olahraga, Seksyen 13, Shah Alam, 40100, Selangor Darul Ehsan, Malaysia
LiveDNA: 60.15110