Research Article
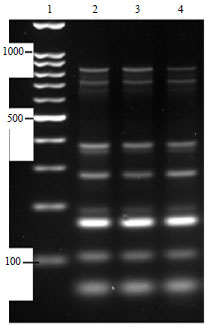
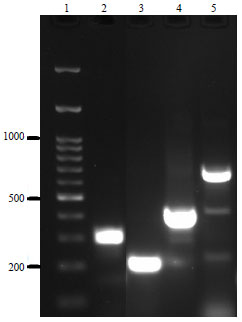
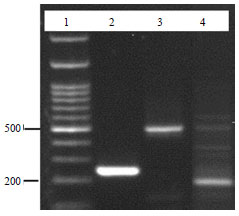
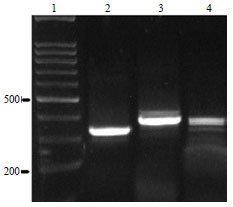
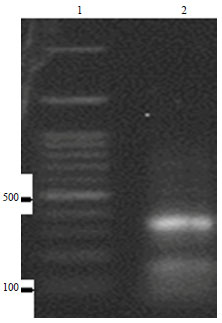
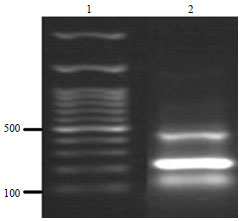
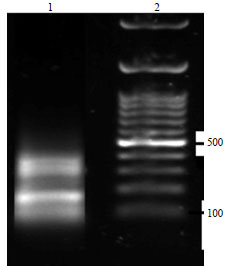
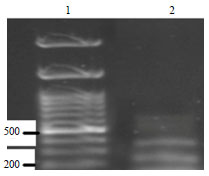
Molecular Characterization of Pasteurella multocida Isolated from Rabbit in Egypt
Division of Genetic Engineering and Biotechnology, Department of Cell Biology, National Research Centre, Egypt
LiveDNA: 20.1134
M.S. Abdel-Salam
Division of Genetic Engineering and Biotechnology, Department of Microbial Genetics, National Research Centre, Egypt
LiveDNA: 20.10464
S.S. Hafez
Department of Botany, Faculty of Women for Arts, Science and Education, Ain Shams University, Egypt
W. Nemr
Department of Radiation Microbiological, National Centre for Radiation Research and Technology, Atomic Energy Authority, Egypt
LiveDNA: 20.12391