Research Article
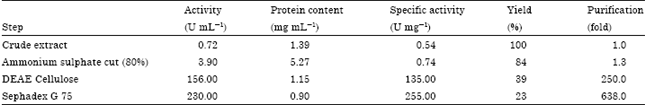
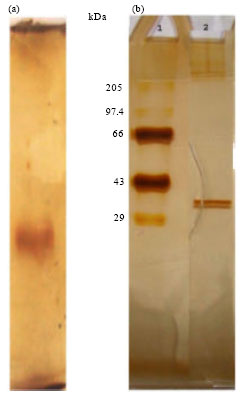
Purification and Properties of Novel Malate Dehydrogenase Isolated from Pseudomonas aeruginosa
Centre for Marine Science and Technology, Manonmaniam Sundaranar University, Rajakkamangalam-629 502, Kanyakumari District, Tamil Nadu, India
Surgen A. Bright
Centre for Marine Science and Technology, Manonmaniam Sundaranar University, Rajakkamangalam-629 502, Kanyakumari District, Tamil Nadu, India
Anuj Nishanth Lipton
Department of Biotechnology, Udaya School of Engineering, Vellamodi, Kanyakumari District, Tamilnadu, India
S.G. Prakash Vincent
International Centre for Nanobiotechnology, Centre for Marine Science and Technology, Manonmaniam Sundaranar University, Rajakkamangalam-629 502, Kanyakumari District, Tamil Nadu, India