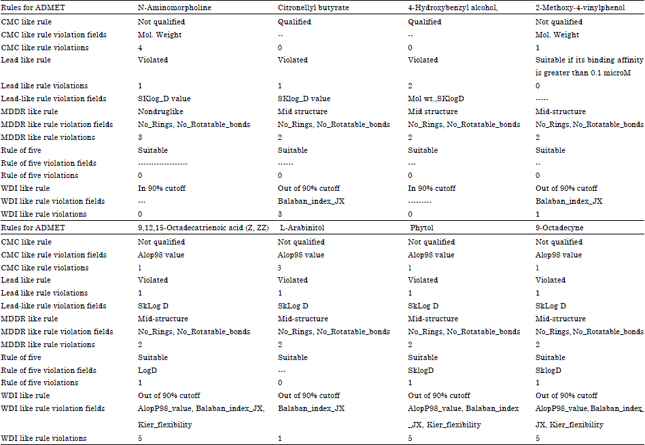
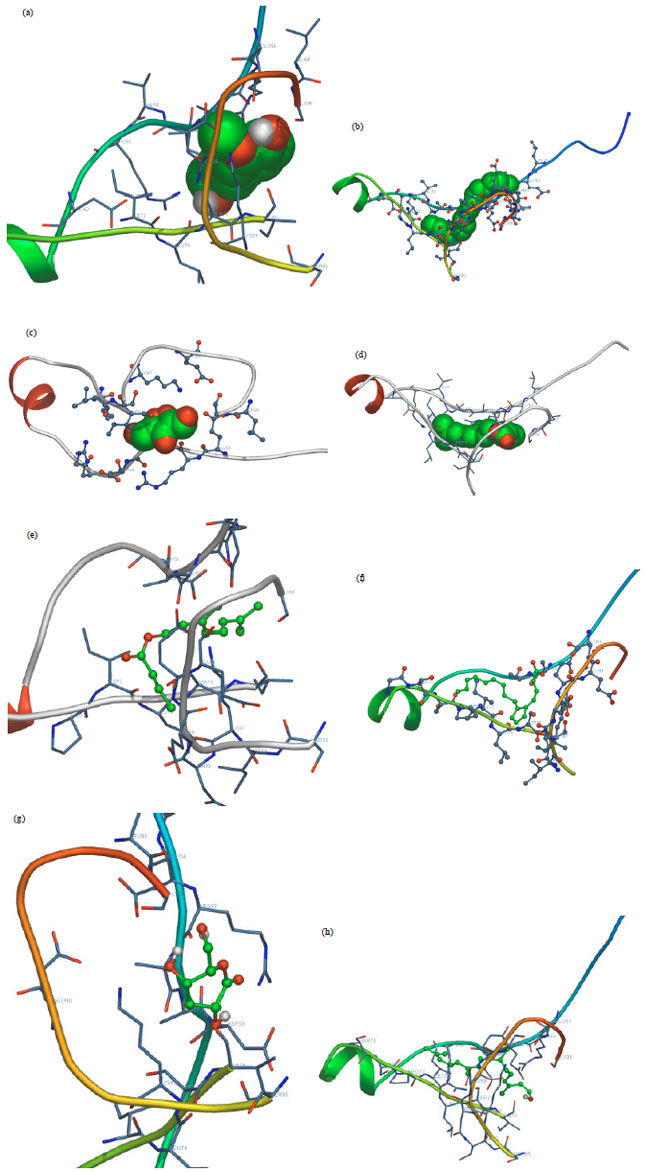
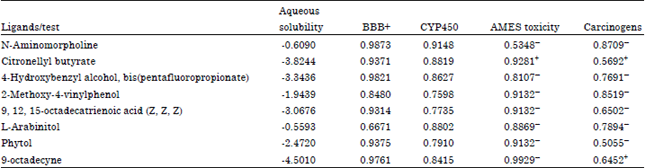
Research Article
Evaluation of Natural Compound as a Potential Drug Against DENV Non-structural Proteins: In silico Study
Centre for Advanced Computing and Bioinformatics, CRD, PRIST University, Vallam, Thanjavur, India
Anusha Bhaskar
Centre for Advanced Computing and Bioinformatics, CRD, PRIST University, Vallam, Thanjavur, India
P. Kumarasamy
Madras Veterinary College, Chennai, India