Research Article
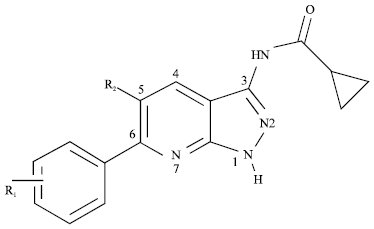
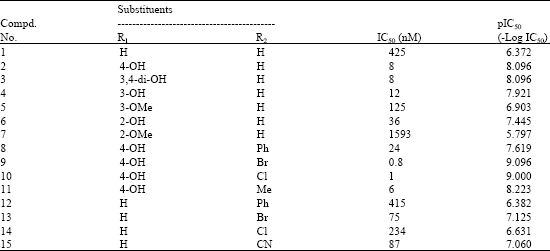
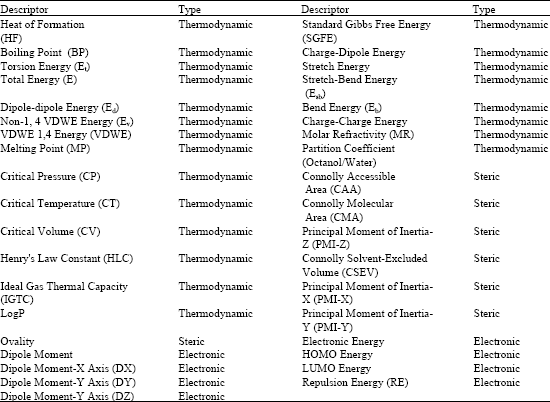
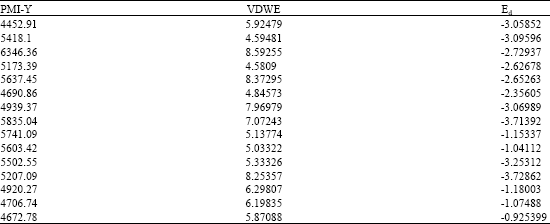
QSAR Studies on Some GSK-3α Inhibitory 6-aryl-pyrazolo (3,4-b) pyridines
Department of Pharmacy, Shri G. S. Institute of Technology and Science, 23 Park Road, Indore-452003, Madhya Pradesh, India
C. Karthikeyan
Department of Pharmacy, Shri G. S. Institute of Technology and Science, 23 Park Road, Indore-452003, Madhya Pradesh, India
S.K. Sharma
Maharaja Surajmal Institute of Pharmacy, New Delhi, India
N.S. Hari Narayana Moorthy
School of Pharmaceutical Sciences, Rajiv Gandhi Proudyogiki Viswavidyalya, Airport Bypass Road, Gandhi Nagar, Bhopal-462036, Madhya Pradesh, India
P. Trivedi
Department of Pharmacy, Shri G. S. Institute of Technology and Science, 23 Park Road, Indore-452003, Madhya Pradesh, India