Mini Review
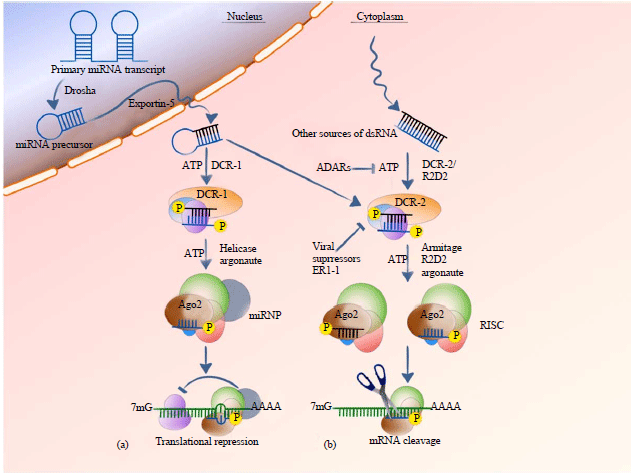
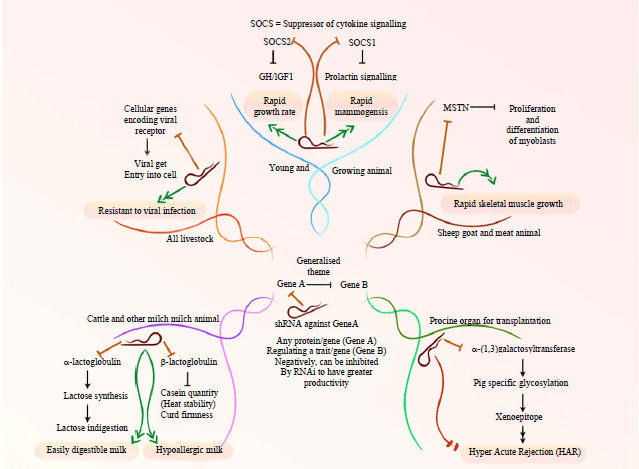
RNAi Mediated Transgenesis for Improving Animal Produce
Division of Veterinary Biotechnology, Indian Veterinary Research Institute, Izatnagar, 243122 Bareilly, Uttar Pradesh, India
Saurabh Gupta
Division of Veterinary Biotechnology, Indian Veterinary Research Institute, Izatnagar, 243122 Bareilly, Uttar Pradesh, India
LiveDNA: 91.10672
Ajay Kumar Singh
Division of Veterinary Biotechnology, Indian Veterinary Research Institute, Izatnagar, 243122 Bareilly, Uttar Pradesh, India
Manish Mahawar
Division of Animal Biochemistry, Indian Veterinary Research Institute, Izatnagar, 243122 Bareilly, Uttar Pradesh, India
Ravikumar Gandham
Division of Veterinary Biotechnology, Indian Veterinary Research Institute, Izatnagar, 243122 Bareilly, Uttar Pradesh, India
Sujoy K. Dhara
Division of Veterinary Biotechnology, Indian Veterinary Research Institute, Izatnagar, 243122 Bareilly, Uttar Pradesh, India
LiveDNA: 91.1986