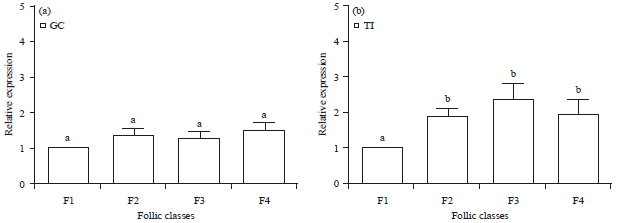
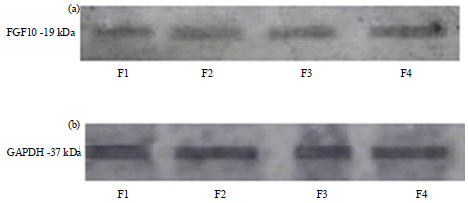
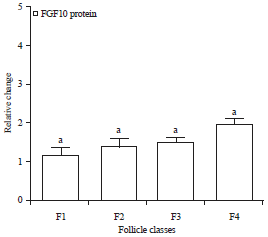
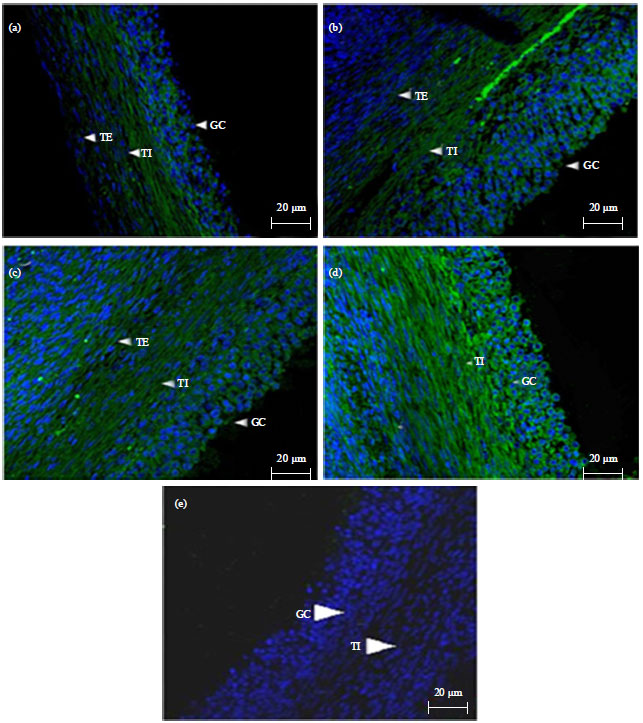
Research Article
Expression and Localization of Fibroblast Growth Factor 10 (FGF10) in Ovarian Follicle During Different Stages Development in Buffalo
Physiology and Climatology, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
Jaya Bharati
Physiology and Climatology, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
M.K. Bharti
Physiology and Climatology, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
G. Singh
Physiology and Climatology, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
M. Sarkar
Physiology and Climatology, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India