Research Article
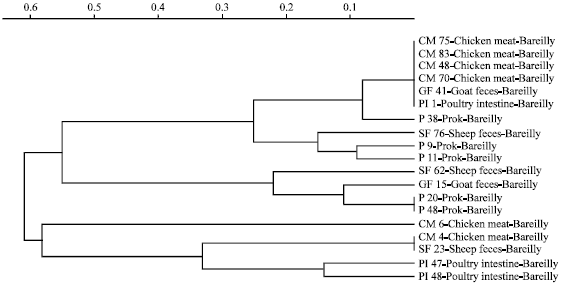
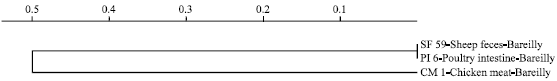
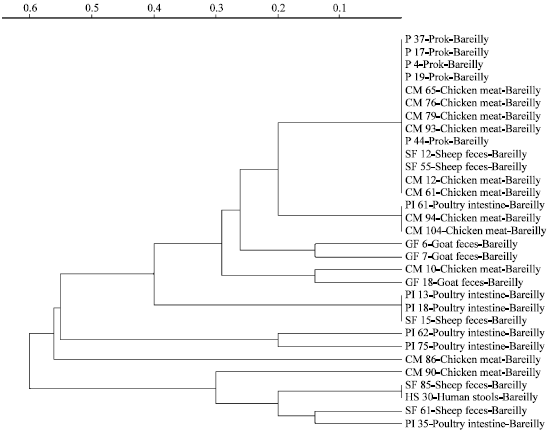
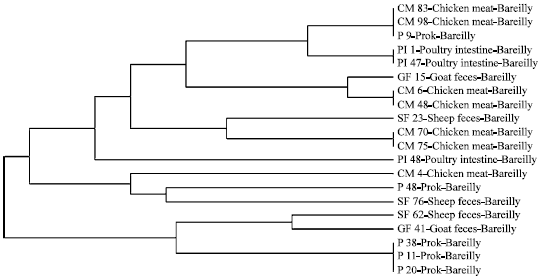
Molecular Characterization of Arcobacter Isolates Using Randomly Amplified Polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR)
Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
R.S. Rathore
Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
T.P. Ramees
Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
H.V. Mohan
Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
M. Sumankumar
Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
R.K. Agarwal
Division of Bacteriology and Mycology, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
A. Kumar
Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India
K. Dhama
Division of Pathology, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, 243122, India