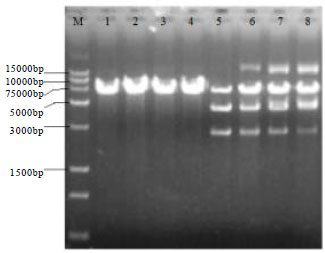
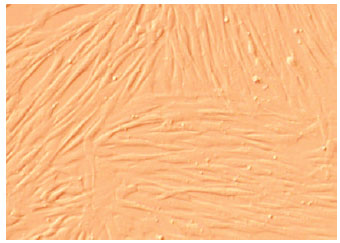
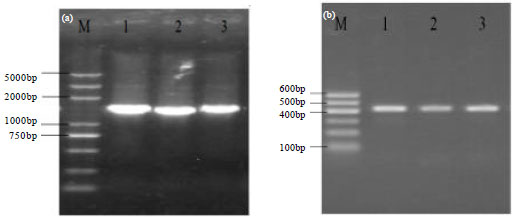
Research Article
Construction of Myostatin Gene Knockout Vector in Goat and Transfection Of Nuclear Donor Cells
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China
Ying-Hui Ling
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China
Chun-Huan Ren
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China
Xiao Cheng
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China
Hong-Guo Cao
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China
Xiao-Fei Guo
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China
Ya-Feng Huang
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China
Xiao-Rong Zhang
College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Peoples Republic of China