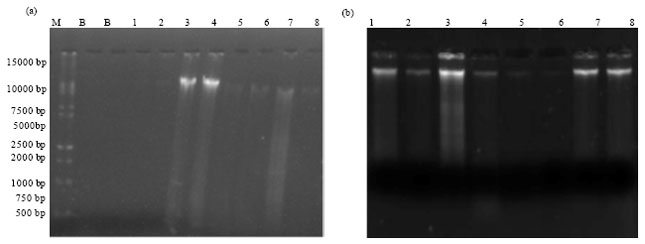
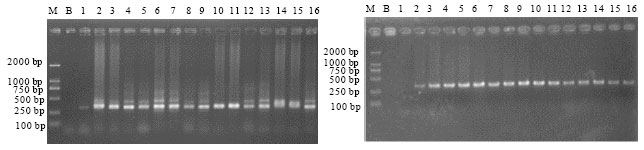
Short Communication
Rapid Salt-Extraction of Genomic DNA from Formalin-Fixed Toad and Frog Tissues for PCR-Based Analyses
Department of Biology Science, Fuyang Normal College, Fuyang City 236041, China
Ke-Jun Zhang
Qingdao Institute of Biomass Energy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao City 266101, China