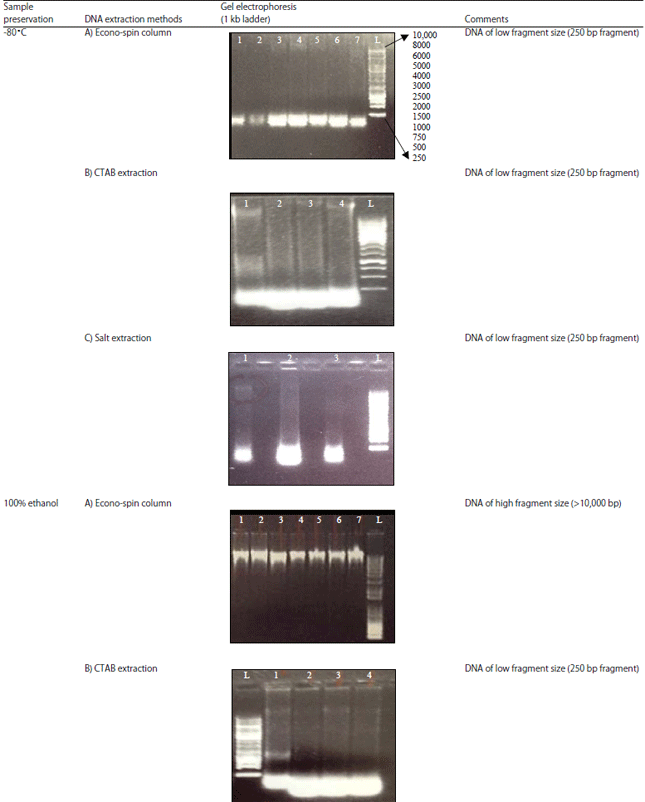
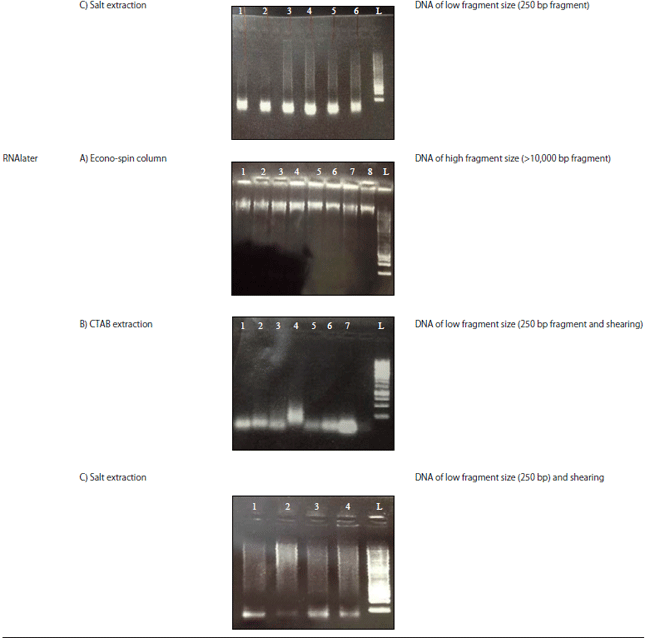
Research Article
Development of an Efficient Method for Producing High Quality Genomic DNA in Crustaceans
Griffith School of Environment, Australian Rivers Institute, Griffith University, 170 Kessels Road, 4111 Nathan, Australia
LiveDNA: 880.16968
Daniel Schmidt
Griffith School of Environment, Australian Rivers Institute, Griffith University, 170 Kessels Road, 4111 Nathan, Australia
Jane Hughes
Griffith School of Environment, Australian Rivers Institute, Griffith University, 170 Kessels Road, 4111 Nathan, Australia