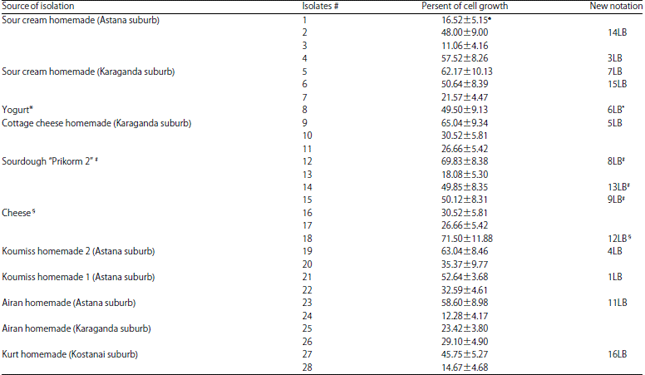
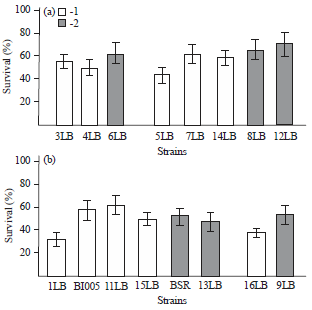
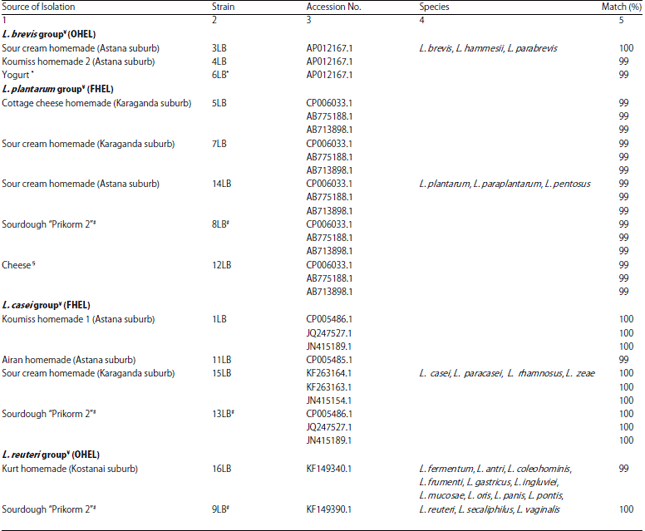
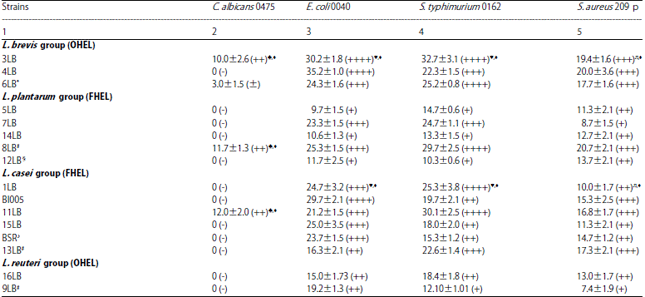
Research Article
Investigation of Acid and Bile Tolerance, Antimicrobial Activity and Antibiotic Resistance of Lactobacillus Strains Isolated from Kazakh Dairy Foods
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
G. Abitayeva
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
S. Anuarbekova
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
D. Shaikhina
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
K. Li
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
S. Shaikhin
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
LiveDNA: 7310.12583
K. Almagambetov
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
A. Abzhalelov
RSE Republican Collection of Microorganisms, SC MES RK, Astana, Kazakhstan
S. Saduakhassova
Nazarbayev University, Astana, Kazakhstan
A. Kushugulova
Nazarbayev University, Astana, Kazakhstan
F. Marotta
ReGenera Research Group for Aging Intervention and Milano Medical, Milano, Italy