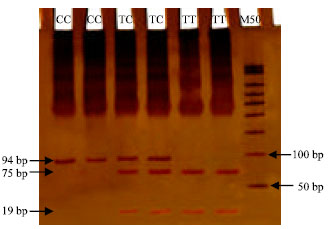
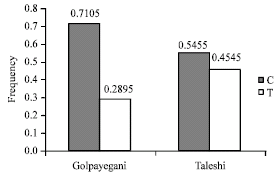
Research Article
Leptin Gene Polymorphism in Iranian Native Golpayegani and Taleshi Cows
Department of Animal Science, College of Agriculture, Ferdowsi University of Mashhad, Mashhad, P.O. Box 91775-1163, Iran
Alireza Heravi Moussavi
Department of Animal Science, College of Agriculture, Ferdowsi University of Mashhad, Mashhad, P.O. Box 91775-1163, Iran
Abdul Raoof Alashawkany
Department of Animal Science, College of Agriculture, Ferdowsi University of Mashhad, Mashhad, P.O. Box 91775-1163, Iran
Shahrokh Ghovati
Department of Animal Science, College of Agriculture, Ferdowsi University of Mashhad, Mashhad, P.O. Box 91775-1163, Iran