Research Article
Evaluation of Antifungal Activity of Purified Chitinase 42 from Trichoderma atroviride PTCC5220
Faculty of Science, Razi University, Iran
M. R. Zamani
National Institute of genetic Engineering and Biotechnology, Tehran, Iran
M. Motallebi
National Institute of genetic Engineering and Biotechnology, Tehran, Iran









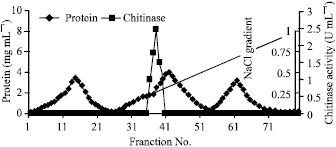
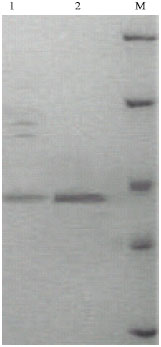
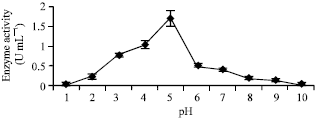
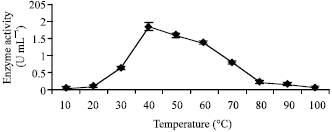
sathiyamoorthy Reply
excellent work and interesting work most of very use for my work guideline.