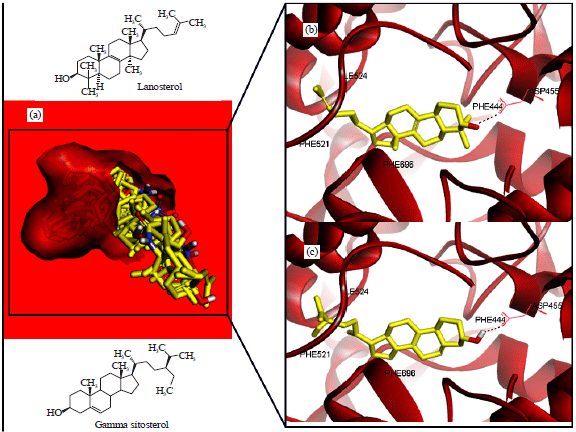
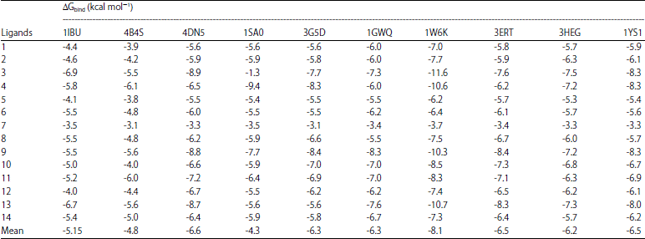
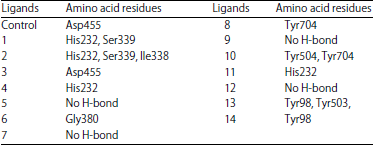
Research Article
Antimicrobial, Antioxidant and T47D Cytotoxic Activities of Trichaptum sp., A Fungal Endophyte from Phyllantus niruri Linn.: In vitro and in silico Studies
Program of Pharmacy, Faculty of Science and Technology, Ma Chung University, 65151 Malang, Indonesia
Maywan Hariono
Faculty of Pharmacy, Sanata Dharma University, Maguwoharjo, Sleman, 55282 Yogyakarta, Indonesia